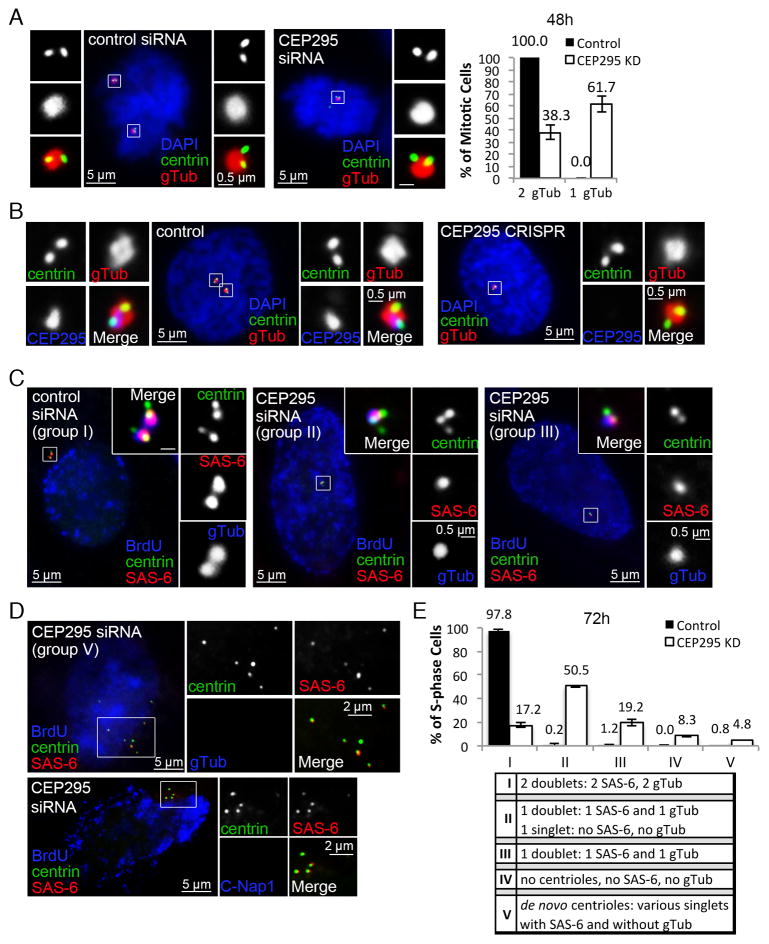
Figure 2. Loss of CEP295 does not block centriole assembly but leads to centrosome reduction.
(A) U2OS cells transfected with control or CEP295 siRNA for 48hr were stained with indicated antibodies. γ-tubulin (gTub) marks active centrosomes. Only mitotic centrosomes were examined (boxed and magnified), and quantified (n>50, N=3, right). Error bars represent standard deviation.
(B) RPE1 cells in which CEP295 gene was mutated by CRISPR/Cas9 gene targeting were analyzed and stained with indicated antibodies. Control cells were nucleofected with Cas9 and empty gRNA vectors. Early prophase cells are shown. ~15% of cells were effectively targeted by CRISPR (not shown).
(C) Centriole duplication was analyzed in control or CEP295-RNAi cells in S phase 72hr after transfection, with indicated antibodies. BrdU was added 30 min before fixation. Unlike control cells carrying two centriole doublets (group I), majority of CEP295-RNAi cells contained only one centriole doublet, with or without an additional centrin singlet devoid of SAS-6 (group II & III).
(D) De novo centrioles were found in a small percentage of CEP295-depleted cells lacking centrosomes (group V), labeled with centrin and SAS-6, but not with γ-tubulin or C-Nap1.
(E) Quantification of centriole duplication (n>160, N=3). Four groups of duplication defects were seen in CEP295-depleted cells as indicated (II–V). Error bars represent standard deviation.
See also Figure S1.