Figure 1.
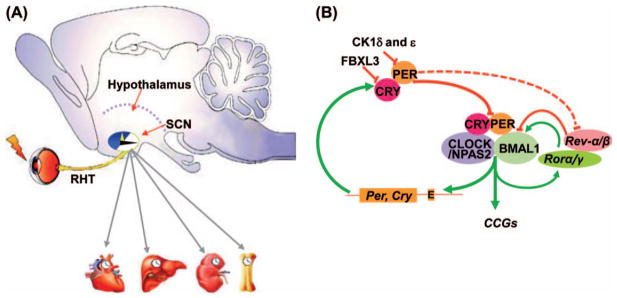
The mammalian circadian clock. (A) The mammalian circadian clock is comprised of circadian input pathways, such as the light input pathway via the retinohypothalamic tract (RHT), the central clock located in the hypothalamic suprachiasmatic nucleus (SCN), the circadian output pathways including the neuroendocrine and autonomic nervous systems (grey arrows) and peripheral clocks in all tissues studied. (B) A simplified model of the molecular clock. Solid lines show direct regulation of the positive and negative feedback loops by core circadian genes Bmal1, Clock, Ck1δ and ε, Cry, Naps2, Per, Rev-erbα and β, as well as Rorα and γ. The dashed lines show indirect regulation of Rev-erbα and β by PER. The molecular clock also targets clock-controlled genes (CCGs) that regulate diverse cellular processes. The first-order CCGs are controlled by the molecular clock directly at the transcriptional level.
