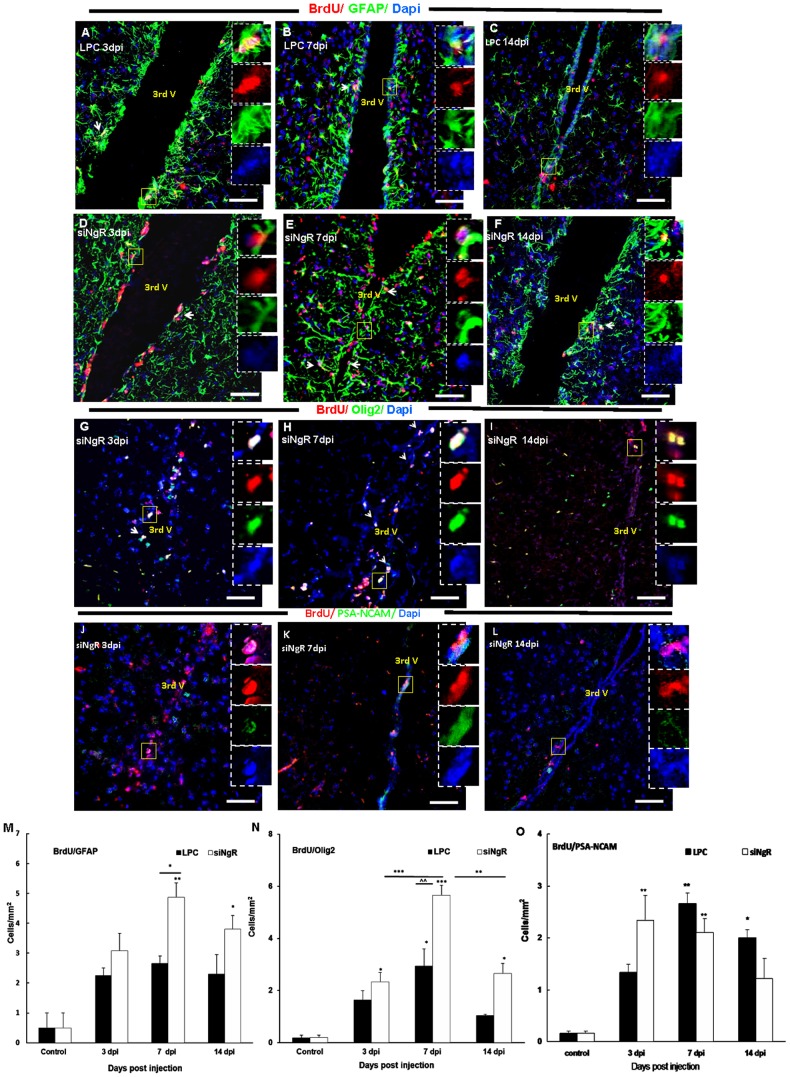
Figure 7. Third ventricle progenitor cell activation in response to LPC-induced lesions in optic chiasm is greater following NgR inhibition.
Double labeling of BrdU (red) with Olig2, GFAP or PSA-NCAM (green) was done in coronal sections of third ventricle area in different groups. (A–C) BrdU+/GFAP+ cells in the rims of third ventricle of LPC group at 3 (A), 7 (B) and 14 dpi (C). (D–F) BrdU+/GFAP+ cells in the rims of third ventricle of LPC+siNgR treated animals at 3 (D), 7 (E) and 14 dpi (F). (G–I) BrdU/Olig2 double staining in third ventricle of LPC+siNgR treated animals at 3 (G), 7 (H) and 14 (I) dpi (see fig. S1D–F for BrdU+/Olig2+ cells in LPC group). (J–L) BrdU/PSA-NCAM positive cells in the third ventricle of LPC+siNgR treated animals at 3 (J), 7 (K) and 14 dpi (L) (see fig. S1G-I for BrdU+/PSA-NCAM+ cells in LPC group). Square shows the cells magnified in inset. Arrows show double marker positive cells. (M–O) Histograms show quantification of double marker positive cells, BrdU+/GAFP+ (M), BrdU+/Olig2+ (N) and BrdU+/PSA-NCAM+ (O) in different groups. Double marker positive cells per area/mm2 are averaged from the counts of nine sections of each brain, n = 3. Statistical analysis used two-way ANOVA with Bonferroni's post-test. Differences between groups were significant (p<0.001). The number of BrdU+/GFAP+ cells in third ventricle of LPC 3, 7 and 14 dpi was increased but changes were not significant compared to Control 7dpi (mice received saline inside chiasm), but this type of cells in LPC+siNgR treated animals at 3, 7 and 14 dpi were considerably increased compared to Control (*p<0.05, ***p<0.001, **p<0.01, respectively) (M). The number of BrdU+/Olig2+ cells was considerably increased in siNgR-LPC group at 3, 7 and 14 dpi compared to Control (**p<0.01, ***p<0.001; respectively) (N). Additionally in these animals, at 7 dpi the number of BrdU+/Olig2+ was considerably increased compared to LPC 7 dpi (∧∧∧p<0.001) (N). The number of BrdU/PSA-NCAM positive cells in the third ventricle of LPC+siNgR treated animals was significantly increased at 3 and 7 dpi (both, p<0.01), while it was significantly increases in LPC treated animals at 7 and 14 dpi (p<0.05, p<0.01; respectively). Data are expressed as Mean ±SEM, Bars: 50 µm.