Abstract
Under conventional conditions, mice deficient in core 1-derived O-glycans (TM-IEC C1galt1−/−), which have a defective mucus layer, experienced spontaneous inflammation of the colon. Analysis of fecal bacterial populations by pyrosequencing of 16S rRNA gene showed that disease in conventional TM-IEC C1galt1−/− was associated with shifts in the microbiota manifested by increases in Lactobacillus and Clostridium species, and decreases in unclassified Ruminococcaceae and Lachnospiraceae. Under germ-free (GF) conditions, TM-IEC C1galt1−/− presented decreased goblet cells, but did not develop inflammation. Monoassociation of GF TM-IEC C1galt1−/− revealed that bacterial species differ significantly in their ability to induce inflammatory changes. Bacteroides thetaiotaomicron caused inflammation, while Lactobacillus johnsonii (enriched during colitis) did not. These observations demonstrate that not all microbiota shifts that correlate with disease contribute to pathogenesis.
Keywords: gut microbiota, pyrosequencing, core 1 O-glycans deficient mice, dysbiosis, spontaneous colitis, Bacteroides thetaiotaomicron, Lactobacillus johnsonii, Bacteroides sartorii, Clostridium, Akkermasia muciniphila
Introduction
Inflammatory bowel diseases (IBD) are a range of phenotypes from Crohn disease (CD) to ulcerative colitis (UC). Most spontaneous rodent models of IBD require the presence of bacteria to develop disease. This requirement has been proved in several models of inflammatory disorders, such as the interleukin 2-deficient mice, the interleukin 10-deficient mice, and the HLA-B27/β2m transgenic rat, in which the host does not develop inflammation if kept germ-free (GF).1
Differences in pathological changes in mucus expression have been found between UC and CD.2-4 While the thickness of the mucus layer and the number of goblet cells either remain unchanged or are increased in CD, both of these elements are reduced in UC.5 Both patterns of change have an impact on the number of bacteria present in the colon, as some constituents of the microbiota live in close association with the mucus layer and contribute to niche establishments for other members of the intestinal microbiota.6 Accordingly, preserving mucus components and the mucus layer is essential in its role as a barrier, by separating the gut microbiota from the intestinal epithelia and mucosal immune cells. We hypothesize that when the mucus layer is impaired by inflammation, genetic loss, or overconsumption by gut bacteria, these changes can result in increased inflammatory signaling and colitis. Defects associated with the mucus layer or its components disrupt the relationship between the microbiota and the host, thus potentially evoking immune responses against the microbiota predisposing to disease. The Core 1 β1,3-galactosyltransferase, C1GALT1 (also known as T-synthase), exclusively controls the formation of the core 1 structure, which can be extended and modified in different ways, such as sialylation, sulfation, and fucosylation. Conventional mice that lack this enzyme specifically in intestinal epithelial cells (IEC C1galt1−/−) spontaneously develop colitis.7 In addition, conventional adult mice with inducible deficiency of core 1 O-glycans in the intestinal epithelium (TM-IEC C1galt1–/–) also develop spontaneous colitis. Broad-spectrum antibiotic treatment reduces colitis in these models, indicating a requirement for intestinal bacteria in colitis development in these mice.
The role of microbes in colitis has long been observed and studied8,9 and the complex interactions of host genetics, microbiota and even viral triggers have been discussed.10 However, no clear role for any specific microbes in UC has been identified. We examined here if the TM-IEC C1galt1−/− mouse model (backcrossed onto the C56BL/6 background) in a GF setting will develop inflammation, and identify the specific microbes that promote colitis in the absence of normal mucus production.
To this end, we rederived TM-IEC C1galt1−/− in a GF setting. GF TM-IEC C1galt1−/− mice were analyzed to determine whether a breach in the mucus barrier in the absence of microbes predisposes to inflammation. Moreover, aiming to characterize changes in the gut microbiota associated with a breach in the mucus layer caused by loss of mucin type O-glycans, we performed molecular analysis of fecal bacterial populations by pyrosequencing of 16S rRNA genes within the intestinal luminal content of conventional TM-IEC C1galt1−/− mice. Furthermore, we performed monoassociation experiments of GF WT (TM-IEC Cre negative C1galt1flox/flox mice used as controls) and GF TM-IEC C1galt1−/− (TM-IEC Cre positive C1galt1flox/flox) mice with selected bacterial species to determine those associated with the development of inflammation in the context of a defective mucus layer.
Results
GF TM-IEC C1galt1−/− mice showed decreased number of goblet cells and did not develop inflammation relative to the conventionalized TM-IEC C1galt1−/− mice
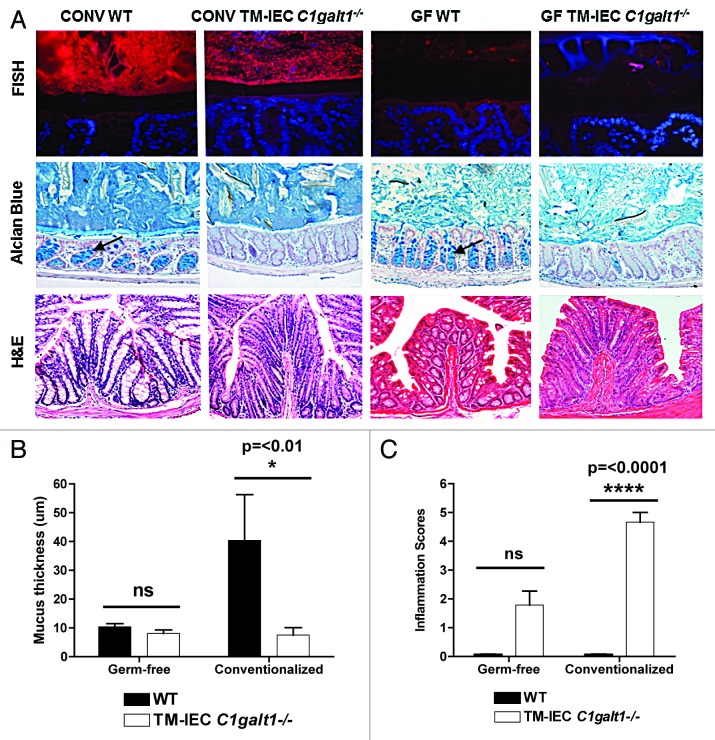
FISH staining of the distal colonic tissue confirmed absence of bacteria in the GF mice, and showed robust bacterial colonization and closer proximity of the microbiota to the epithelial cells in the conventionalized TM-IEC C1galt1−/− mice (Fig. 1A). Both Tn antigen staining and Alcian Blue (AB) staining were used to assess deletion-induced mucus loss. Tn antigen represents the unmodified substrate for C1GALT1 and is therefore increased when C1GALT1 activity is absent. As expected, GF and conventionalized TM-IEC C1galt1−/− mice showed an increased presence of Tn antigen compared with their WT counterparts (Fig. S1A). AB-stained colon tissue of conventionalized TM-IEC C1galt1−/− mice and GF TM-IEC C1galt1−/− mice showed decreased number of goblet cells and a thinner mucus layer relative to their WT counterparts (Fig. 1A). It is known that GF mice have a thinner mucus layer than conventional mice.11 Interestingly, upon deletion induction, the mucus layer thickness decreased further from an average of 10.4 microns in the GF WT to 8.1 microns in the TM-IEC C1galt1−/− mice. In the conventionalized mice, the mucus layer decreased from an average of 40.4 microns to 7.6 microns (P < 0.01) in the WT and TM-IEC C1galt1−/− mice, respectively (Fig. 1B), thus confirming closer proximity of the microbial communities to the epithelium in the TM-IEC C1galt1−/− mice. Specifically for the GF setting, counting of goblet cells in the TM-IEC C1galt1−/− mice showed a statistically significant decrease of approximately 25% (P = < 0.05) in the number of goblets cells when compared with GF WT mice (data not shown).
Figure 1. Germ-free TM IEC C1galt1−/− mice did not develop inflammation. (A) FISH staining with universal probes demonstrated robust colonization (bacteria stained in red) of the conventionalized (ex-germ-free) mice. Mucosa-associated bacteria increased in the conventionalized TM-IEC C1galt1−/− mice. Gene deletion induced a decrease in the mucus layer (black space, between bacteria -in red- and epithelial cells -in blue) relative to the WT control. Alcian Blue staining showed a decrease in mucus production by goblet cells (black arrows point at goblet cells in the WT mice). H&E was used to assess epithelial hyperplasia in the colon. Conventionalized TM-IEC C1galt1−/− developed a high degree of epithelial hyperplasia. Differences in the appearance of the tissue and length of the crypts were observed in the conventionalized TM-IEC C1galt1−/− mice compared with the GF counterpart and the WT controls. (B) Data represents the mean thickness of the inner mucus layer obtained from 3 to 13 animals for each genotype from both, conventionalized and germ-free mice. Statistics were determined by 1-Way ANOVA with Bonferonni’s post-test. Error bars = SEM (C) GF TM-IEC C1galt1−/− mice did not develop inflammation relative to conventionalized TM-IEC C1galt1−/− mice based on the established parameters. For (B) and (C), error bars represent standard deviations. P values were estimated for changes between genotypes (WT vs. TM-IEC C1galt1−/−).
Clinical disease signs of colitis, such as development of diarrhea, weight loss, and rectal prolapse were absent or not significant (average daily weight gain and standard deviations per treatment group and per genotype are listed in Table S1). Classic colitis scores include loss of goblet cells, but since loss of core 1-derived O-glycans decreases the size and number of goblet cells, we used a modified inflammation scoring system (Fig. S1B) based on infiltration of immune cells into the colonic lamina propria and epithelial hyperplasia evaluated by H&E staining (Fig. 1A). Inflammation was further addressed by immunolabeling of CD45+ cells (Fig. S1A and S1C). We observed low levels of inflammation in the GF TM-IEC C1galt1−/− mice that could be explained by an increased exposure of the intestinal epithelia (thus increased stimuli of immune cells) to environmental and food antigens as a consequence of a thinner mucus layer, as it has been shown that even chemically-defined ultrafiltered “antigen-free” diets elicit immune responses.12 However, statistical analysis of inflammation scores showed no significant differences between the genotypes in the GF setting (GF WT vs. GF TM-IEC C1galt1−/− mice). In contrast, the conventionalized TM-IEC C1galt1−/− mice developed moderate to severe inflammation, relative to the conventionalized WT counterpart, with scores values of 4 and 5 in a scale with a highest possible value of 6 (P < 0.001) (Fig. 1C).
Distinct microbial communities are associated with the conventional colitic TM-IEC C1galt1−/− mice
To determine whether lack of core 1 O-glycans caused proportional changes in the gut microbiota of the mice, we analyzed fecal pellets, cecal contents, and left (distal) colon contents of conventional TM-IEC C1galt1−/− mice (n = 4) and conventional wild type (WT) mice (n = 4) by pyrosequencing of the V1-V2 region of the 16S rRNA gene. It is important to note that the results from the three anatomical sites are different, suggesting that exclusive analysis of fecal microbiota may have limitations. We focused our analysis on the colon contents, the site of inflammation. A total of 406,339 reads were obtained, which, after submission to quality control parameters and chimera removal, resulted in 279 912 sequences (average length of 294 base pairs). One sample (fecal pellets from wild type mouse) was dropped from the analysis because of poor amplification of reads. The percentage of chimeric sequences removed was 16.4%.
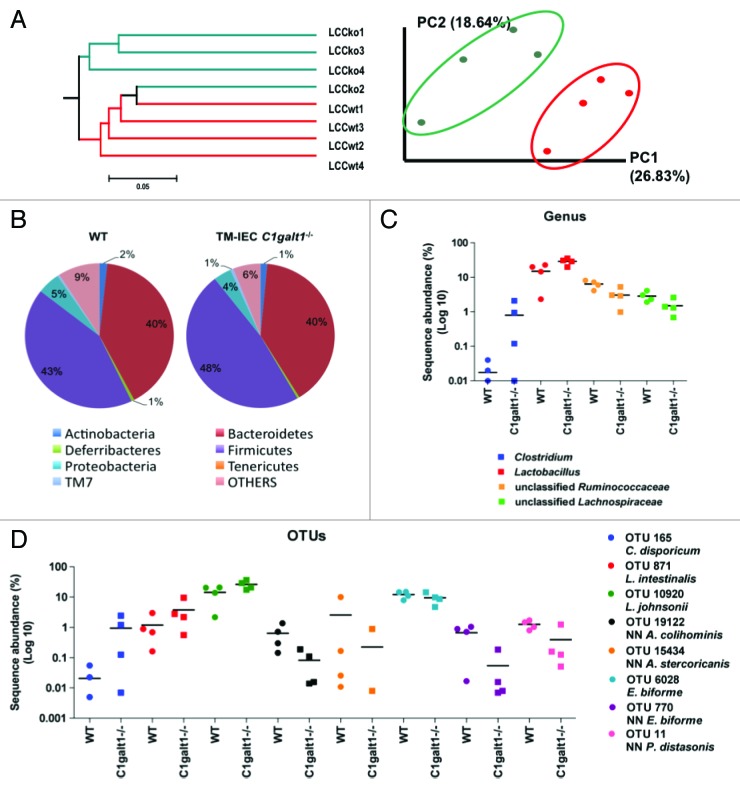
Unweighted UniFrac is a metric for distance between communities of organisms that takes into consideration absence or presence of Operational Taxonomic Units (OTUs), but not their abundance.13 Principal Component Analysis (PCA) is a variable reduction procedure, done with the objective of eliminate redundancy or variable correlation with one another. Unweighted UniFrac and PCA were performed to assess differences in microbial composition between the samples. Analyses showed differences in bacterial communities between the genotypes TM-IEC C1galt1−/− vs. WT, specifically in the left colon content. Principal component 1 accounts for 26.83% of the variability in the data, and principal component 2 accounts for 18.6% in the left colon content (Fig. 2A). The UniFrac phylogenetic tree showed a distinct microbial population exclusively in the left colon content of the conventional TM-IEC C1galt1−/− mice when compared with the conventional WT (Fig. 2A). These differences were not seen in the fecal pellets or cecal contents of the mice (Fig. S2A and B).
Figure 2. Deficiency of Core 1 O-glycans is associated with microbial changes that are the net result of increases in Clostridium and Lactobacillus species and decreases in unclassified Lachnospiraceae, unclassified Prevotellaceae and unclassified Ruminococcaceae in the mice gut. (A) Unweighted UniFrac (non-abundance) cluster tree and PCA graph, performed to assess similarities in microbial communities between the genotypes, revealed two distinct microbial communities in left colon content (LCC) samples. KO: TM-IEC C1galt1−/− mice. Red dots and lines represent the WT mice. Green dots and lines represent the TM-IEC C1galt1−/− mice. (B) Lack of core 1 O-glycans caused increases in Firmicutes in the LCC at the phylum level. (C) Increases in Firmicutes are the net result of statistically significant increases in Clostridium and Lactobacillus population and decreases in unclassified Ruminocccaceae. (D) Statistical analyses of OTUs showed changes in 8 OTUs specific to the LCC of the TM-IEC C1galt1−/− mice. These OTUs are represented by Clostridium disporicum, Lactobacillus intestinalis, Lactobacillus johnsonii, which are increased in the LCC of the mice; and Anaerotruncus colihominis, Allobaculum stercoricanis, Eubacterium biforme and Parabacteroides distasonis, which are decreased in the mice colon. For (C) and (D), horizontal lines represent means. n = 3–4 for each anatomical site tested, data points equal to zeros are not shown as the y-axis is in log10 scale.
Sequences were analyzed using the Ribosomal Database Project (RDP) Classifier (http://rdp.cme.msu.edu).14,15 The total of sequences per taxa per sample was summarized at different taxonomic levels, and proportions were calculated. Significance of the Classifier data was determined as mentioned in the methods section, using a critical value of 3.29 (α = 0.001) to assess significance of results.
RDP-Classifier-based analysis at the phylum level showed a significant increase in the number of Firmicutes in the TM-IEC C1galt1−/− relative to the WT mice (Fig. 2B; Fig. S3A). Firmicutes in left colon content samples increased from 42.6% in the WT mice to 47.7% in the TM-IEC C1galt1−/− mice (Z = 4.12). Results at the family level are discussed in Figures S4 and S5.
At the genus level, significant changes were found in left colon content samples on populations of Lactobacillus, Clostridium, unclassified Lachnospiraceae, and unclassified Ruminococcus (Fig. 2C). The predominant change was in the population of Lactobacillus, which increased from 14.26% to 28.67% (Z = 14.29). Clostridium also increased changing from 0.01% to 0.89% (Z = 5.56). Changes in these two groups of bacteria were found in all anatomical sites (Fig. 2C; Fig. S3B and C). Unclassified Lachnospiraceae diminished in left colon content samples from 6.23% to 3.03% (Z = -6.00). Unclassified Ruminococcaceae decreased in all types of samples reaching statistical significance in the left colon contents (from 2.87% to 1.49%; Z = -3.7391).
One hundred and eighty-four (184) unique OTUs were used for analysis, which accounted for 97.6% of the sequences. Eight OTUs with significant changes were identified in the left colon contents samples (Fig. 2D). Three OTUs, representing clusters associated with Lactobacillus intestinalis (97% identity), Lactobacillus johnsonii (99% identity), and Clostridium disporicum (97% identity), were increased in the conventional TM-IEC C1galt1−/− mice. The number of Lactobacillus johnsonii doubled in left colon contents from 13.60% to 25.79% (Z = 12.34), while Lactobacillus intestinalis increased almost 3-fold from 1.29% to 3.77% (Z = 6.43). Clostridium disporicum increased from less than 0.02% in conventional WT mice samples to 1.08% (Z = 6.01) in the conventional TM-IEC C1galt1−/− mice.
The remaining OTUs that were found significant in the distal colon were reduced in the conventional TM-IEC C1galt1−/− mice relative to the conventional WT, some of them having over a 10-fold reduction. These were: OTU 11, whose nearest neighbor (NN) shares 94% identity with Parabacteroides distasonis, reduced from 1.34% to 0.45% (Z = -3.6872) and OTU 19122 NN (96% identity to Anaerotruncus colihominis), from 0.73% to 0.09% (Z = -3.8169). Additionally, three OTUs from the family of Erysipelotrichaceae were reduced: Eubacterium biforme (97% identity), from 12.66 to 9.35% (Z = -4.18); OTU 770 NN (94% identity to Eubacterium biforme), from 0.61% to 0.06% (Z = -3.6155); and OTU 15434 (91% identity to Allobaculum stercoricanis) from 2.98% to 0.28% (Z = -8.1531). Proportions and Z-values per anatomical site for all OTUs are given in Table S2.
Additionally, pyrosequencing analysis of cecal contents from conventionalized WT and TM-IEC C1galt1−/− mice showed differences in the microbial communities between the genotypes. RDP Classifier analysis of sequences obtained from the conventionalized mice showed increases in the Clostridiaceae I in the TM-IEC C1galt1−/− mice relative to the WT (from 4.32% to 12.59%, Z = 8.77), which agrees with what was found for the conventional group. However, opposite to the conventional mice group, the Lactobacillaceae decreased in the conventionalized TM-IEC C1galt1−/− mice when compared with the WT (from 22.49% to 11.63%, Z = -8.43) (Fig. S7 and S8).
Development of inflammation is microbe specific
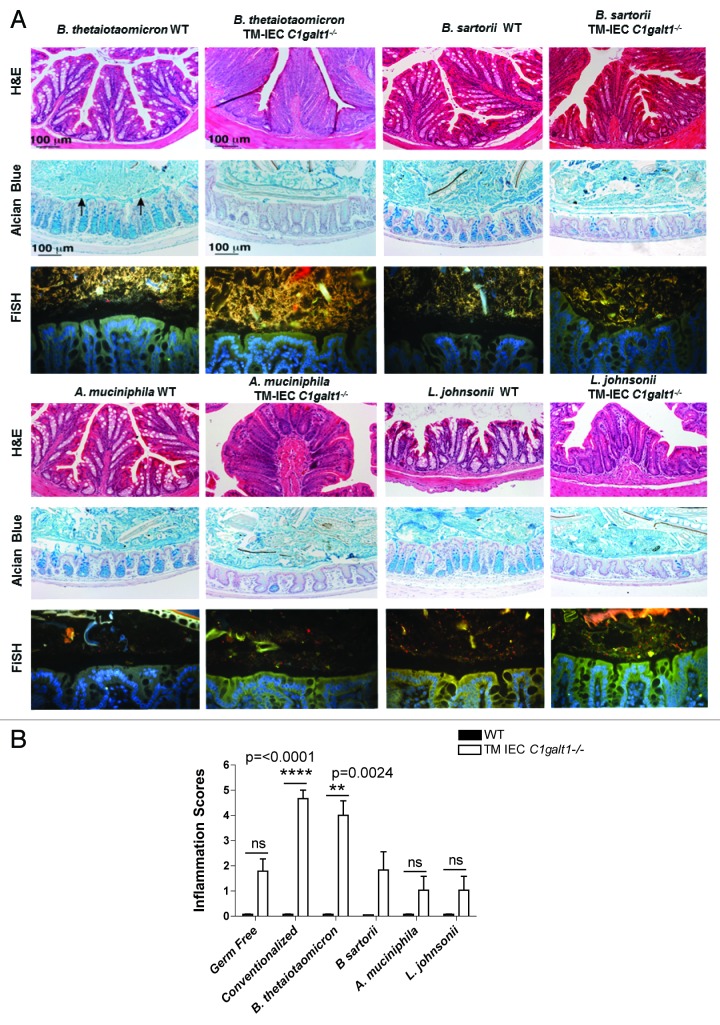
In this initial characterization of gnotobiotic mice we performed monoassociation experiments of GF TM-IEC C1galt1−/− mice with selected bacterial species. Bacteroides thetaiotaomicron is the best-characterized gut bacteroides to date, with previous studies showing that this bacterium can induce colitis.16,17 In addition, it is known to directly interact and digest mucus.18 Likewise, we tested Akkermansia muciniphila, another gut microbe associated with mucus degradation that has been implicated in both protection as well as provocation of IBD.19,20 Moreover, we took advantage of the fact that one of the major microbes significantly altered in the microbiota of conventional TM-IEC C1galt1−/− mice was Lactobacillus johnsonii, an easily cultivated organism isolated by culturing bacteria from feces of colitic mice. Finally, we tested an alternative Bacteroides, Bacteroides sartorii, as a mouse specific Bacteroides, as it has been observed that host adaptation by gut microbes can have significant impact on in vivo fitness.21 Therefore, we predicted that mouse specific autochthonous bacteria, such as B. sartorii, may have a different effect on colitis development compared with the allochthonous B. thetaiotaomicron.
The highest inflammatory scores (determined as describe above) were obtained for the conventionalized TM-IEC C1galt1−/− mice (P < 0.0001), followed by monoassociation of TM-IEC C1galt1−/− mice with B. thetaiotaomicron (P = 0.0024) relative to their WT counterparts (Fig. 3B). Monoassociation with B. thetaiotaomicron caused the highest degree of infiltration of LP CD45+ cells (P = 0.0173) into the colon tissue among the bacterial treatments (Fig. S6) and the highest degree of hyperplasia (P = 0.0323) among the monoassociation experiments (Fig. 3A). Interestingly, the inflammation scores obtained for mice monoassociated with B. sartorii, A. muciniphila, and L. johnsonii were equivalent or lower than the ones calculated for GF TM-IEC C1galt1−/− mice (Fig. 3B). Interestingly, the conventionalized mice did not develop the expected level of inflammation (described by Fu et al.).7 This is most likely the consequence of differences in composition between the resident microbiota of the conventional mice and the donor microbiota (addressed in Fig. S7) used for this experiment. These differences could arise from environmental and handling protocols that are distinct from one facility to the other, among other factors. For example, the conventional mice microbiota includes member of the Helicobacteraceae. This family was not found in the cecum of the conventionalized mice (colonized with Peterson laboratory mice microbiota, which had been monitored and kept helicobacteria free). Even though changes in the levels of this species did not reach statistical significance between the genotypes, the presence of Helicobacter species has been associated with susceptibility to inflammation in some murine models of colitis.22 In contrast, Allobaculum spp. comprise an average of 19% of the gut microbiota in the conventionalized mice, while this bacterial genus was not identified in the original conventional mice. Knowledge on the role and metabolic capabilities of Allobaculum is limited. Its effect in inflammation is unknown, thus opening to the possibility of a role in disease prevention.
Figure 3. Intestinal symbiont Bacteroides thetaiotaomicron is capable of inducing colitis in the absence of core 1 O-glycans. (A) Assessment of epithelial hyperplasia in WT and TM-IEC C1galt1−/− monocolonized mice showed differences in the appearance of the tissue between the genotypes (monoassociated WT vs monoassociated TM-IEC C1galt1−/−) caused by increases in volume as a consequence of increases in the number of epithelial cells. AB staining revealed an impaired mucus production by the goblet cells (“empty” crypts, lack of blue stain observed in the TM IEC C1galt1−/− mice). FISH analysis revealed robust colonization by Bacteroides spp. in both WT and TM-IEC C1galt1−/− mice, and moderate or weak colonization by the other bacteria tested. (B) The TM-IEC C1galt1−/− conventionalized mice (with a complete microbiota) presented the highest degree of hyperplasia among all bacterial treatments. Monoassociation with B. thetaiotaomicron caused the highest degree of hyperplasia among the monoassociation experiments. Bars represent fold change over WT; error bars represent standard deviations. P values were estimated for changes between genotypes (WT vs. TM-IEC C1galt1−/−).
Discussion
The role of the gut microbiota in IBD is easy to envision, widely studied, and yet remains enigmatic. There are many mouse models of IBD, and most are dependent on the gut microbiota. The TM-IEC C1galt1−/− mice provide us with a model of IBD that has a defined molecular defect, as opposed to complicated and poorly understood immunological mechanisms such as cytokine knockouts, or HLA-B27 transgene expression. The goal of our studies has been to use this well-defined defect (lack of core-1 derived O-glycans in the intestinal tract) to identify the changes it incites in the microbiota, and how different microbes vary in their potential to induce inflammation. Ultimately, our results reinforced the observation that microbiota profiling alone will not reveal the pathways to IBD pathogenesis.
We observed that the severe disease seen in conventionally raised mice did not develop in GF and even ex-GF (conventionalized) mice. GF mice are known to have a distinct intestinal morphology and an altered immune system, a lower number of goblet cells and a thinner mucus layer in the small intestine than conventional WT mice.23 TM-IEC C1galt1−/− mice did not develop disease as long as they were kept germ-free, but showed histological signs of inflammation after being conventionalized through gavage of conventional mouse cecal contents. Therefore, it is clear in this model that the microbiota is necessary for disease development.
Changes between microbial communities were found in the conventional TM-IEC C1galt1−/− mice relative to the conventional control (WT) mice. Summarizing, these changes mainly include increases in Firmicutes spp. and reductions in Bacteroidetes spp. in intestinal epithelial specific core 1 O-glycan-deficient mice. Bacteria use the mucus layer for attachment and niche establishment,24 thus a thinner mucus layer in the TM-IEC C1galt1−/− mice could explain dysbiosis. Moreover, altered glycan profiles themselves will likely alter community composition by modifying the repertoire of glycan structures that serve as attachment sites or energy sources that assist in specific tropism and establishment of commensal species. Furthermore, since some bacteria are capable of inducing inflammation,25 the development of inflammation in the colon, and other related immune changes (i.e., induction of proinflammatory cytokines and cell-mediated responses) could affect niche establishments for some members of the microbiota. Uncoupling the role of changes in O-glycosylation vs. inflammation in promoting dysbiosis in this model is challenging, as these two facets are intimately linked due to impaired mucus barrier in the absence of core 1 O-glycans.
It is very interesting that monoassociation with B. thetaiotaomicron induced inflammation similar to conventionalization, while colonization with B. sartorii did not. Both organisms are Bacteroides, capable of colonizing the host to similar levels, and are metabolically similar in their ability to grow on glycan substrates as observed by Benjdia et al.26 (and data not shown). However, differences between these bacteria could arise in the expression of different forms of LPS or other TLR agonist/antagonist that could impact inflammation in mice,27 physiological differences in carbohydrate utilization (and even digestion of the remaining mucus), and/or production of anti-inflammatory metabolites, such as short-chain fatty acids, that have been implicated in suppressing inflammation. Experimental evidence shows that gut microbes have evolved into host-adapted lineages through long-term evolutionary processes.21 B. sartorii (and L. johnsonii) used in these set of experiments were mice isolates. The low intensity of inflammation induced in the TM-IEC C1galt1−/− mice by these two strains suggests that strain-host evolution could play a role in reducing the pathogenic potential of different bacterial species in this mice model.
The L. johnsonii strain used in the monoassociation experiments was isolated from conventional colitic TM-IEC C1galt1−/− mice, where it was increased in proportion of the microbiota relative to conventional WT mice. However, the TM-IEC C1galt1−/− mice mono-colonized with this bacterium did not have significant inflammation. Our results show parallel agreement with Bloom et al.16 who found that an Enterobacteriaceae isolate failed to cause disease in antibiotic-pretreated mice even though their population was highly enriched in mice who developed spontaneous disease, thus supporting that not all gut microbial changes are causative of disease. As research into the microbiota of IBD continues to identify changes associated with disease, our results suggest that increased microbes should not be considered causative, but indeed could be compensatory, or even beneficial.
Our results show that after a breach in the mucus barrier, IBD progresses through specific changes in the constitution of the microbiota. The similarities between the human disease and the results found in our mutant mice model—namely increases in Lactobacillus28 and Clostridium29 and decreases in Lachnospiraceae30—highlight the value of the TM-IEC C1galt1−/− mouse model. The control afforded by this model in terms of timing of the introduction of the host’s defect (a decreased intestinal mucosal layer), and in terms of the extent of microbial colonization of the intestines of GF mice (monoassociation vs. conventionalization) have been instrumental in the elucidation of the complex interplay of factors involved with provoking inflammation in the context of a failed mucus barrier.
Clearly, specific components of the microbiota play critical roles within the gut, either in prevention or development of inflammation. Accordingly, alterations in the epithelial barrier and the mucus layer that in turn affect the microbiota could either be beneficial (i.e., increasing microbes like L. johnsonii) or detrimental (i.e., increasing Bacteroides and decreasing Lachnospiraceae) to the host. Advances in our understanding of mucosal immunology and of the composition, structure and metabolic profile of the gut microbiome will help elucidate the role of each player in the development of inflammation and of appropriate therapies. Fecal transplant and other manipulations of the microbiota may become part of normal treatment of UC.31 Future studies with individual microbes in our model will help to refine UC treatment goals, and understand the mechanisms that underlie the successes and failures of these empiric approaches.
Materials and Methods
Germ-free TM-IEC C1galt1−/− mice
Breeding of C1galt1−/− mice with VillinCre-ERT2 transgenic mice, both on a C57BL/6J congenic background, generated Tamoxifen (TM)-inducible intestinal epithelial cell-specific C1galt1−/− mice (TM-IEC C1galt1−/−) which were made germ-free (GF) at Taconic Farms, Inc. Mice were bred in the Gnotobiotic Mice Facility at the University of Nebraska-Lincoln and weaned at four weeks of age. Mice were genotyped as established by Fu et al.7 Six to ten mice, six to seven weeks old were used per experiment including Cre-negative (WT), age-matched littermates as controls. Mice were colonized by either oral gavage of 200 μl of cecal contents previously harvested from two conventional C57BL/6J mice and diluted in 4 ml of sterile PBS, gavaging 200 μl of single pure bacterial cultures, or by rubbing the culture onto the coat of each mouse.32 Tamoxifen (TM; MP Biomedicals #0215673980) was injected intraperitoneally (1mg in an ethanol/sunflower oil mixture 1:9 v/v) for five consecutive days in a blinded fashion to both, WT (Cre-) and TM IEC C1galt1−/− (Cre+) mice. TM treatment was started 48 to 72 h post-colonization in both GF and colonized WT and TM IEC C1galt1−/− mice. Mouse weight was monitored and recorded every other day. They were sacrificed 20 or 30 d after TM injections. We first analyzed 20 d (as the time with significant inflammation was observed in conventionally raised mice) and extended to 30 d when no clinical signs or symptoms were observed. Details and a schematic representation of the experimental protocol are provided in Figure S9.
Sample collection
For mice experiments, mice were anesthetized using isofluorane (Baxter #1001936060) and blood collected for immunoglobulin measurement. Cecum contents were snap-frozen in liquid nitrogen. The distal colon was divided into three sections and preserved in liquid nitrogen, Carnoy’s fixative (60% methanol, 30% chloroform, 10% glacial acetic acid), or 10% formalin, respectively.
For pyrosequencing, samples for the conventional data set were collected from four conventional wild type mice, and four conventional TM-IEC C1galt1−/− mice kept in a specific pathogen-free facility at the Oklahoma Medical Research Foundation (OMRF), Oklahoma City, Oklahoma. Samples included fecal pellets (FP), cecal contents (CC) and left (distal) colon contents (LCC), and they were kept at -80 °C until processed for DNA isolation. Samples for the conventionalized data set were collected from four wild type mice and four conventionalized TM-IEC C1galt1−/− mice (colonized as previously described) kept in the Gnotobiotic Mice Facility at the University of Nebraska in Lincoln, Nebraska. DNA was isolated as previously described by Martinez et al.33 with minor modifications. Details are provided in Supplemental Materials.
Immunohistochemical and immunofluorescence staining of the distal colon tissue
Preparation of samples and procedures for Hematoxylin and Eosin (H&E) staining (to address epithelial hyperplasia), Alcian Blue (AB), and Tn antigen staining (to assess deletion induction), CD45+ cells staining (for evaluation of inflammatory processes), and FISH protocol (to assess bacterial association) are discussed in SI. H&E and AB were applied as established in the Core Imaging Facility of the Oklahoma Medical Research Foundation (OMRF) (http://imaging.omrf.org/wp-content/uploads/2011/08/hande.pdf. Tn antigen was stained as described by Fu et al.7
Data analysis for determination of inflammation scores and mucus thickness
CD45+ LP cells were stained as described in SI and counted in a blinded fashion at 40× magnification, for 4 to 8 fields per sample, by counting all the nuclei in between the crypts, avoiding the nuclei associated with blood and lymphatic vessels (i.e., endothelial cells). Calculation of the number of epithelial nuclei in the crypts, by counting 10 to 20 well-oriented crypts per section using the H&E stained slides, assessed epithelial hyperplasia. The average number of intra-epithelial cells (IECs) per crypt for each section was obtained from the raw number of IECs. The average number of LP cells and IECs for each TM IEC C1galt1−/− mouse was divided by the average number obtained for each WT, and converted to percentage. Scores from zero to six were assigned (three for CD45+ cells count and three for epithelial hyperplasia) considering changes relative to WT controls (Fig. S1B). Statistical analysis between genotypes was determined by unpaired t test with Welch’s correction using GraphPad Prism 5.0 (GraphPad Software Inc) statistical software.
For determination of mucus thickness using AB-stained tissue sections, ten to 50 measurements were taken from each animal colon preserved in Carnoy’s fixative (over 1–3 10× fields per section, 1 or 2 sections per animal), depending on the quality of mucus layer preservation. Measurements were taken at equal intervals (70 μm apart) established by a grid overlaying the image. Each measurement was taken perpendicular to the upper and lower borders of the inner mucus layer, which are clearly delineated when this layer is intact and well preserved. ImageJ software was used for data acquisition. Statistics were determined by 1-Way ANOVA with Bonferonni’s post-test using GraphPad Prism 5.0 (GraphPad Software Inc) statistical software.
Pyrosequencing of 16S rRNA PCR and data analysis
For the conventional mice, the variable regions 1 and 2 of the 16s rRNA genes were amplified by PCR using a composite forward primer and a reverse primer containing a unique 12-base barcode to tag PCR products to their respective samples. For the conventionalized mice, primers against the variable regions 3 to 5 were used along with unique 10-base barcodes attached to the reverse primer. Primers and protocol are described in Supplemental Materials. Sequences generated from pyrosequencing were submitted to the open source software package Quantitative Insights into Microbial Ecology (QIIME; http//qiime.source-forge.net) for removal of low quality sequences. Sequences that met the following parameters were kept for analysis: (1) length between 264 and 324 nucleotides for the conventional mice data set, and between 510 and 570 for the conventionalized mice data set; (2) no ambiguous bases; (3) quality score equal or above 25; (4) no mismatches in primers; and (5) no mismatches in barcodes. After sequences were quality controlled, chimera check was performed using the Chimera Slayer method in QIIME and binned by barcode. Taxonomic-based analysis was performed using the Classifier13 program of the Ribosomal Database Project (RDP) version 9 (http://rdp.cme.msu.edu/)14 to assign taxonomic status to each sequence. Significance of the Classifier data was determined by calculating proportions and statistical analysis performed by calculating the average mean of proportions and Z score (formula discussed in Supplemental Materials) using a critical value of 3.29 (α = 0.001).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We would like to thank Jorge Torres for his help and support during the realization of the monoassociation experiments, and Ines Martinez for helpful discussions regarding analysis of pyrosequencing data. This work was possible due to funds provided by grants NIAID R21AI097419, K08AI07660901, and R01DK085691. The authors declared no conflict of interest with respect to this manuscript.
Glossary
Abbreviations:
- IBD
inflammatory bowel diseases
- UC
ulcerative colitis
- CD
Crohn disease
- C1GALT1
core 1 β1,3-galactosyltransferase enzyme (also known as T-synthase) which exclusively controls the formation of the core 1 structure in the mucins that form the mucus layer
- IEC C1galt1−/−
mice that lack the C1GALT1 enzyme specifically in the intestinal epithelial cells
- HLA-B27
human leukocyte antigen B27
- TM
tamoxifen
- TM-IEC C1galt1−/− mice
Tamoxifen-induced deficiency of the C1GALT1 enzyme specifically in the intestinal epithelial cells, thus mice are deficient in core 1-derived O-glycans
- GF
germ-free
- WT
wild type mice
- FISH
flourescence in-situ hybridization
- AB
Alcian Blue staining
- H&E
Hematoxylin and Eosin staining
- RDP
Ribosomal Database Project
- QIIME
Quantitative Insights into Microbial Ecology
- PCA
Principal Component Analysis
- OTU
operational taxonomic unit
- LP
lamina propria
References
- 1.Mizoguchi A. Animal models of inflammatory bowel disease. Prog Mol Biol Transl Sci. 2012;105:263–320. doi: 10.1016/B978-0-12-394596-9.00009-3. [DOI] [PubMed] [Google Scholar]
- 2.Gersemann M, Becker S, Kübler I, Koslowski M, Wang G, Herrlinger KR, Griger J, Fritz P, Fellermann K, Schwab M, et al. Differences in goblet cell differentiation between Crohn’s disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 3.McGuckin MA, Eri R, Simms LA, Florin THJ, Radford-Smith G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100–13. doi: 10.1002/ibd.20539. [DOI] [PubMed] [Google Scholar]
- 4.Larsson JM, Karlsson H, Crespo JG, Johansson ME, Eklund L, Sjövall H, Hansson GC. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm Bowel Dis. 2011;17:2299–307. doi: 10.1002/ibd.21625. [DOI] [PubMed] [Google Scholar]
- 5.Strugala V, Dettmar PW, Pearson JP. Thickness and continuity of the adherent colonic mucus barrier in active and quiescent ulcerative colitis and Crohn’s disease. Int J Clin Pract. 2008;62:762–9. doi: 10.1111/j.1742-1241.2007.01665.x. [DOI] [PubMed] [Google Scholar]
- 6.Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–8. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, Wei B, Wen T, Johansson MEV, Liu X, Bradford E, Thomsson KA, McGee S, Mansour L, Tong M, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–66. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 10.Peterson DA, Turnbaugh PJ. A microbe-dependent viral key to Crohn’s box. Sci Transl Med. 2010;2:43ps39. doi: 10.1126/scitranslmed.3001422. [DOI] [PubMed] [Google Scholar]
- 11.Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G327–33. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooijkaas H, Benner R, Pleasants JR, Wostmann BS. Isotypes and specificities of immunoglobulins produced by germ-free mice fed chemically defined ultrafiltered “antigen-free” diet. Eur J Immunol. 1984;14:1127–30. doi: 10.1002/eji.1830141212. [DOI] [PubMed] [Google Scholar]
- 13.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [PMID: 19004872] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom SM, Bijanki VN, Nava GM, Sun L, Malvin NP, Donermeyer DL, Dunne WM, Jr., Allen PM, Stappenbeck TS. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen JJ, Huang Y, Peterson DA, Goeser L, Fan TJ, Chang EB, Sartor RB. The colitis-associated transcriptional profile of commensal Bacteroides thetaiotaomicron enhances adaptive immune responses to a bacterial antigen. PLoS One. 2012;7:e42645. doi: 10.1371/journal.pone.0042645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–9. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 19.Vigsnaes LK, van den Abbeele P, Sulek K, Frandsen HL, Steenholdt C, Brynskov J, Vermeiren J, van de Wiele T, Licht TR. Microbiotas from UC patients display altered metabolism and reduced ability of LAB to colonize mucus. Sci Rep. 2013;3:1110. doi: 10.1038/srep01110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang CS, Ban M, Choi EJ, Moon HG, Jeon JS, Kim DK, Park SK, Jeon SG, Roh TY, Myung SJ, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J. 2010;4:377–87. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- 22.Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, et al. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One. 2013;8:e70783. doi: 10.1371/journal.pone.0070783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson GR, Trexler PC. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut. 1971;12:230–5. doi: 10.1136/gut.12.3.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mackenzie DA, Jeffers F, Parker ML, Vibert-Vallet A, Bongaerts RJ, Roos S, Walter J, Juge N. Strain-specific diversity of mucus-binding proteins in the adhesion and aggregation properties of Lactobacillus reuteri. Microbiology. 2010;156:3368–78. doi: 10.1099/mic.0.043265-0. [DOI] [PubMed] [Google Scholar]
- 25.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253–7. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benjdia A, Martens EC, Gordon JI, Berteau O. Sulfatases and a radical S-adenosyl-L-methionine (AdoMet) enzyme are key for mucosal foraging and fitness of the prominent human gut symbiont, Bacteroides thetaiotaomicron. J Biol Chem. 2011;286:25973–82. doi: 10.1074/jbc.M111.228841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG, Pettersson S, Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 28.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, Järnerot G, Tysk C, Jansson JK, Engstrand L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–, e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 29.Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481–7. doi: 10.1097/01.MIB.0000159663.62651.4f. [DOI] [PubMed] [Google Scholar]
- 30.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, Berry D. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol. 2013;108:1620–30. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]
- 32.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–3. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 33.Martínez I, Wallace G, Zhang C, Legge R, Benson AK, Carr TP, Moriyama EN, Walter J. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–84. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.