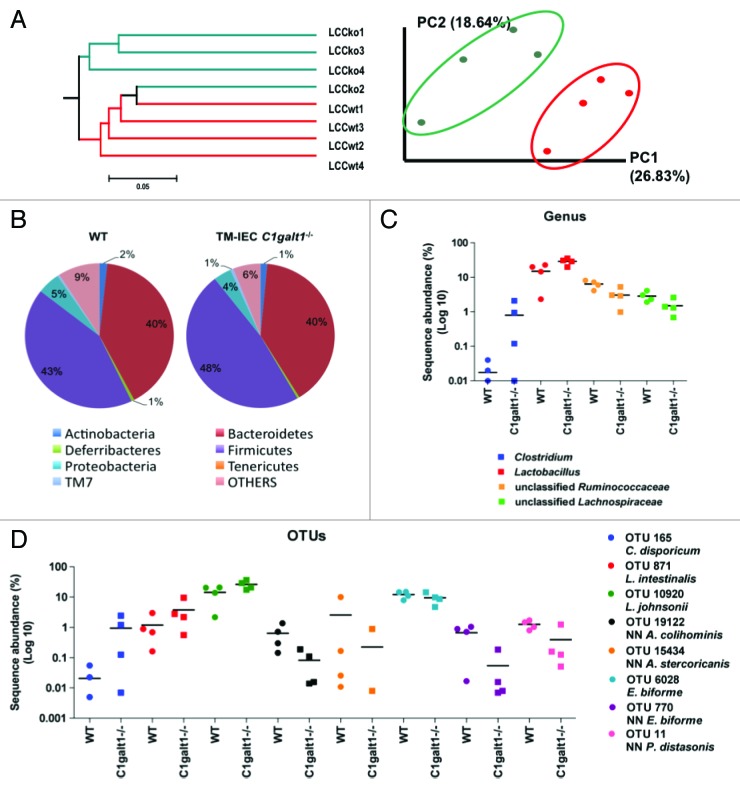
Figure 2. Deficiency of Core 1 O-glycans is associated with microbial changes that are the net result of increases in Clostridium and Lactobacillus species and decreases in unclassified Lachnospiraceae, unclassified Prevotellaceae and unclassified Ruminococcaceae in the mice gut. (A) Unweighted UniFrac (non-abundance) cluster tree and PCA graph, performed to assess similarities in microbial communities between the genotypes, revealed two distinct microbial communities in left colon content (LCC) samples. KO: TM-IEC C1galt1−/− mice. Red dots and lines represent the WT mice. Green dots and lines represent the TM-IEC C1galt1−/− mice. (B) Lack of core 1 O-glycans caused increases in Firmicutes in the LCC at the phylum level. (C) Increases in Firmicutes are the net result of statistically significant increases in Clostridium and Lactobacillus population and decreases in unclassified Ruminocccaceae. (D) Statistical analyses of OTUs showed changes in 8 OTUs specific to the LCC of the TM-IEC C1galt1−/− mice. These OTUs are represented by Clostridium disporicum, Lactobacillus intestinalis, Lactobacillus johnsonii, which are increased in the LCC of the mice; and Anaerotruncus colihominis, Allobaculum stercoricanis, Eubacterium biforme and Parabacteroides distasonis, which are decreased in the mice colon. For (C) and (D), horizontal lines represent means. n = 3–4 for each anatomical site tested, data points equal to zeros are not shown as the y-axis is in log10 scale.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.