Fig. 4. Genetic Mechanism of DNA Methylation and Histone Modification.
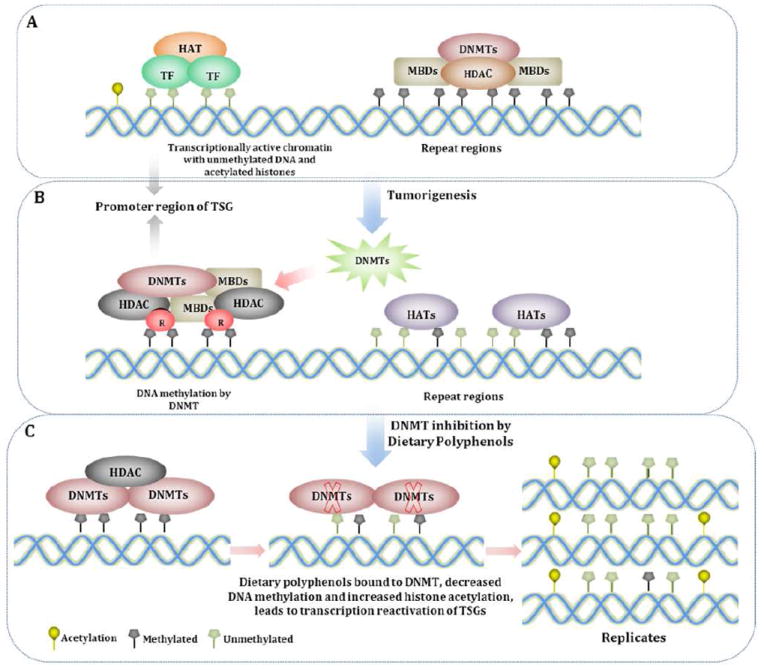
A. Transcriptionally active chromatin with unmethylated DNA and acetylated histones. Genes are unmethylated and packaged with acetylated histone proteins associated with HAT and basal transcription factor machinery. These epigenetic elements constitute an ‘open’ chromatin structure which favors transcription. B. DNA methylation by DNMT. The methylated CpG sites are recognized by the methyl-binding proteins (MBDs), which are associated with repressor (R) and histone deacetyltransferase (HDAC) proteins to remove the acetyl group from the histones, generating a tightly closed chromatin status to shut down gene expression. C. Effects of dietary polyphenols on inhibition of DNMT. DNMT activity is blocked by dietary polyphenols by forming hydrogen bonds with amino acids (Pro, Glu, Cys, Ser, and Arg) in the catalytic pocket of DNMT. Newly synthesized DNA strands are semi-methylated after the first round of DNA replication and become progressively more demethylated after several rounds of replication due to the dilution effect.
