Table 2.
Candidate Arabidopsis TA proteins containing a putative OMM dibasic targeting signal motifa.
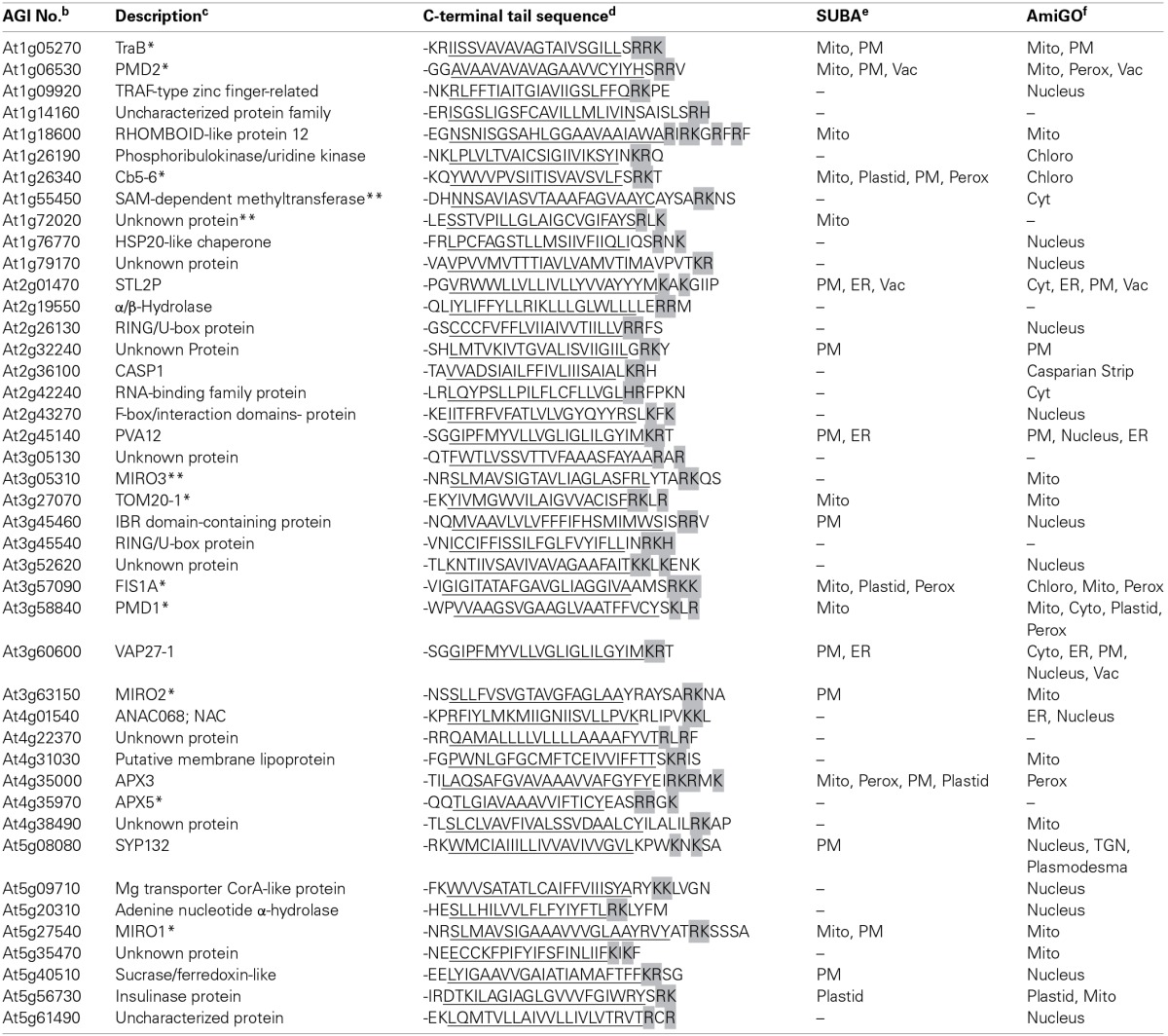
All of the proteins listed are known or predicted to possess a TA orientation (Kriechbaumer et al., 2009; Pedrazzini, 2009; Dhanoa et al., 2010) and also contain a C-terminal mitochondrial dibasic targeting signal motif according to the results of the mutational analysis of selected TA proteins presented this study (see Figures 5–7). See text, including Materials and Methods, for additional details.
bAGI number represents the systematic designation given to each locus, gene, and its corresponding protein product(s) in TAIR. Proteins are listed in ascending order based on their AGI number.
cCommon nomenclature of Arabidopsis TA proteins based on published literature and TAIR. Proteins indicated with an asterisk are also listed in Table 1; proteins indicated with two asterisks were experimentally characterized in terms of their intracellular localization and, for At1g55450, also TA orientation, membrane integration, and C-terminal-tail-dependent targeting; see Figure 8 and text for additional details.
dShown for each protein is its deduced C-terminal tail sequence, including its putative TMD (underlined), based on TOPCONS and visual inspection, and its downstream CTS. Shaded are the dibasic amino acid residues within the mitochondrial dibasic targeting signal motif (i.e., -R/K/H-X{0,1≠E}-R/K/H{≠-H-H- or -H-X-H-}-X{0,1≠E}X{0,3}{CTS=3,8}) found in the CTS of all the proteins shown.
eShown for each protein is its intracellular localization(s) based on published proteomics and/or GFP localization results presented at SUBA3 (The SUBcellular localization database for Arabidopsis proteins) (Tanz et al., 2013). Abbreviations: ER, endoplasmic reticulum; Mito, mitochondria; Perox, peroxisome; PM, plasma membrane; Vac, vacuole.
fShown for each protein is “cellular component” ontology, i.e., where a gene product is located in is a subcompartment of a particular cellular component, based on the AmiGO search tool at the Gene Ontology database (http://www.geneontology.org) (Ashburner et al., 2000). Abbreviations: Cyt, cytosol; TGN, trans-Golgi network; see also above (footnote e).
