Figure 1.
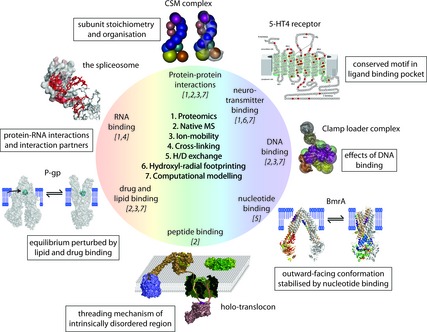
Structural MS and its various techniques. A multitude of structural MS techniques 1 have been applied and combined to study protein–ligand complexes – most of these have been coupled with computational modelling 7. Some recent examples are shown. The subunit stoichiometry and organisation of the CSM complex have been obtained from proteomics and IM‐MS of the intact complex. A model compatible with the available EM density map could thus be obtained [81]. Hydroxyl radical footprinting gave insights into neurotransmitter binding to the GPCR 5‐HT4. The schematic shows the GPCR with amino acids that were studied highlighted in red 33. The clamp loader complex was studied with IM‐MS and computational modelling, allowing construction of a three‐dimensional model of the fully assembled complex 59. Hydrogen–deuterium exchange was applied to study the effect of nucleotide binding on the conformation of the BmrA transporter. Hotspots for hydrogen‐deuterium exchange are represented by colour coding from cold to hot (blue < green < yellow < red) 25. Peptide binding probed by MS uncovered the threading mechanism through the bacterial OmpF 71. IM‐MS gave insights into specific lipid and drug binding to the P‐gp transporter 61. RNA–protein interactions in various spliceosomal complexes have been determined by combining UV‐crosslinking and proteomics. The model derived for the U1‐snRNA (red) in complex with U1‐specific proteins (grey space‐filling) is shown.
