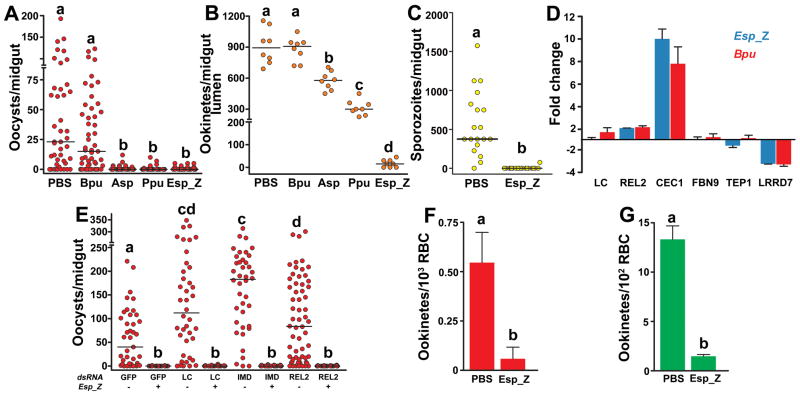
Figure 1. Field bacteria-mediated inhibition of Plasmodium development.
P. falciparum oocyst (A) and ookinete (B) loads in midguts of A. gambiae mosquitoes co-fed with parasites and field-isolated bacteria. Bpu, Bacillus pumilus; Asp, Acinetobacter sp.; Ppu, Pseudomonas putida; Esp_Z, Enterobacter sp. (C) P. falciparum sporozoite loads in salivary glands of A. gambiae mosquitoes co-fed with parasites and Esp_Z. For (A–C), circles represent the number of parasites from an individual mosquito and horizontal lines indicate the median number of parasites per tissue. (D) Midgut-specific transcript abundance of select genes at 8 h after bloodfeeding with equal quantities of either the Bpu or Esp_Z isolate. Each column and error bar represents the fold-change ± standard deviation in transcript abundance when compared to PBS-fed controls. LC= PGRP-LC; CEC1=cecropin1; FBN9=fibrinogen immunolection 9; TEP1=thioester-containing protein 1; LRRD7=leucine-rich repeat-containing protein 7. (E) Oocyst loads in mosquitoes depleted of transcripts for IMD pathway molecules and co-challenged with Esp_Z and P. falciparum. The dsRNA and absence (−) or presence (+) of Esp_Z are indicated below each column. Circles represent the same as in (A). (F–G) In vitro development of P. falciparum (E) and P. berghei (F) ookinetes co-cultured with Esp_Z bacteria. Bars represent the mean ± standard deviation in ookinetes. For all figures, statistical significance is represented by letters above each column, with different letters signifying distinct statistical groups (p<0.05; Mann-Whitney test for (A–C, E); p<0.05; unpaired t-test for (F–G)).