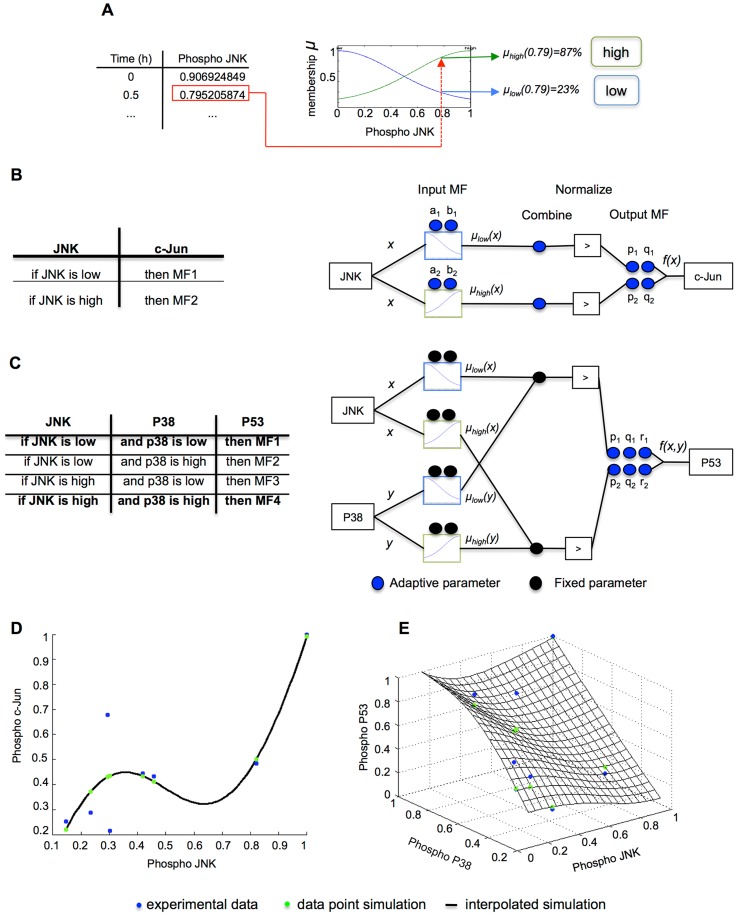
Figure 3. Parameter reduction strategy and data-derived model implementation.
(A) JNK experimental data was transformed into fuzzy sets via 2 Gaussian input membership functions (MF), thereby enabling use of fuzzy logic rules. (B) The table shows logic rules mapping the phosphorylation of JNK to the phosphorylation of c-Jun, as an example. The diagram shows the parameters needed to implement those rules. 8 free parameters are required for the MFs (blue circles), which could increase for other types of function. In addition, the MFs to be combined in each rule need to be determined, here represented as 2 additional free parameters to a total of 10 in this simple FL system with two rules. (C) Implementation of a more complex FL model of P53 regulation by both P38 and JNK to illustrate the parameter reduction strategy. The number of consequent parameters depends on the number of rules, which in turn depend on the number of inputs, hence the setup described in (B) yields here 4 rules (table) with 20 parameters. To reduce the number of free parameters, we fixed the number of input-output combinations (left, in bold), reducing the system to 2 rules and 14 parameters (diagram). By fixing the premise parameters, the number of parameters to estimate was reduced to 6, below the number of experimental time points. (D) Simulation of the model shown in (B). The free parameters were reduced and estimated, and they proved to be sufficient to capture the data trend (blue dots) upon simulation (green dots, RMSE = 0.181), here illustrated in DMSO. The black curve shows the simulation of the model at 100 interpolated data points distributed uniformly. (E) Simulation of the model shown in (C), reduced and trained to the phosphorylation profiles of P38, JNK and P53. The simulation of P53 phosphorylation is depicted here as a mesh (RMSE = 0.223). Training data points are depicted in blue and simulation at the same data point in green. The black surface shows the simulation of the model at 100 interpolated data points distributed uniformly.