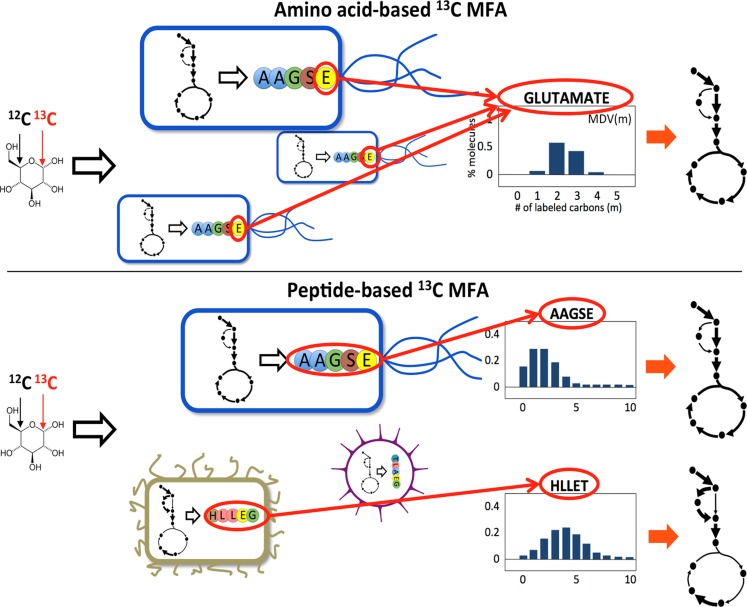
Figure 1. Overview of the traditional amino acid-based and the proposed peptide-based 13C MFA.
For pure (top) and mixed cultures (bottom). A labeled carbon source is provided and the fluxes are derived from the ensuing amino acid labeling profiles. In the case of traditional 13C MFA for pure cultures, contributions from all cells, which are assumed to undergo similar metabolic activities, add up to produce the measured labeling profile (or MDV(m) = fraction of molecules with m 13C atoms incorporated). This measured labeling distribution is then used for the fit (see fig. 2). In the peptide-based method it is the peptide labeling distribution that is used for the fit (see fig. 2). Since peptides are composed of amino acids, its labeling distribution can be easily computed form the amino acid labeling profile, as shown in equation 13. The advantage of this approach is that the peptides can be separated and assigned to different species through standard proteomics techniques.