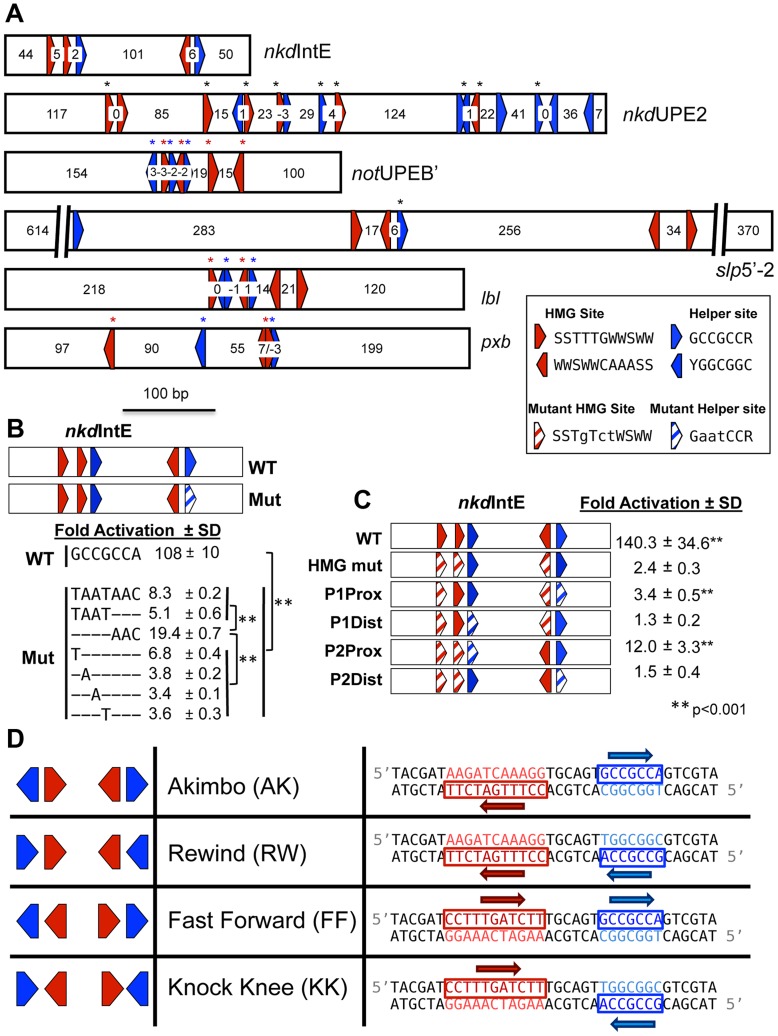
Figure 1. HMG and Helper site configurations in W-CRMs.
(A) Previously characterized W-CRMs [44], [57] with location of predicted HMG (red arrows) and Helper (blue arrows) indicated (see Figure S3 for PWMs of these motifs). Cutoffs for HMG and Helper sites were 5.35 and 6.5, respectively. Direction of arrow indicates orientation of motif (see inset for consensus sequences in both orientations). Number of nucleotides between each motif is indicated. Black asterisks indicate sites that contributed to W-CRM activation by Wnt signaling in cell culture when mutated individually [44], [57]. Red and blue asterisks denote function when all indicated HMG or Helper sites were mutated simultaneously. (B) Systematic mutagenesis of second Helper site in the nkdIntE W-CRM reporter indicates all seven positions contribute to W-CRM activation. Letters indicate mutated nucleotides, with dashes denoting the wild-type sequence. Reporter constructs were transfected into Drosophila Kc cells with or without a plasmid expressing Arm*. Fold activation represents the ratio of Arm*/control, SD equals standard deviation of three biological replicates. (C) HMG and Helper sites work in closely spaced pairs. The nkdIntE reporter contains three functional HMG and two functional Helper sites [44], which were mutated (striped arrows) in different combinations, and tested for Arm* activation in Kc cells. Values represent the mean of three biological replicates ± SD. A Student's T-test was employed to determine significance for data in panels B and C. (D) Nomenclature and symbology for the four possible HMG-Helper site pair orientations. A right pointing arrow indicates the consensus sequence is read 5′ to 3′ on the “top” strand, a left arrow indicates the consensus is read 5′ to 3′ on the “bottom” strand. The sequences shown for HMG and Helper sites are identical to the ones used for DNA binding experiments in Figure 2 and synthetic Wnt reporter constructs (Figure 3A, 4 and 5).