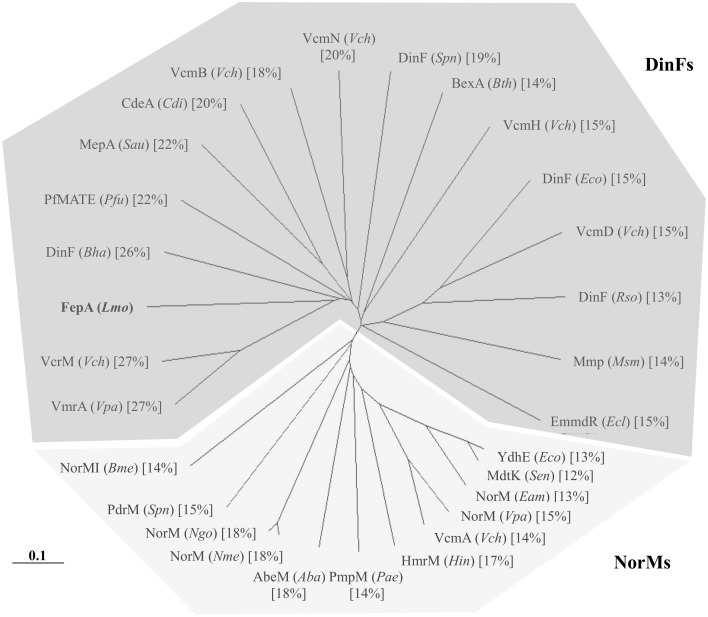
Figure 3. Phylogenetic tree based on neighbor-joining analysis of sequences of bacterial efflux proteins belonging to the MATE family.
The various homologs were identified in: Aba, Acinetobacter baumannii; Bha, Bacillus halodurans; Bme, Brucella melitensis; Bth, Bacteroides thetaiotaomicron; Cdi, Clostridium difficile; Eam, Erwinia amylovora; Ecl, Enterobacter cloacae; Eco, Escherichia coli; Hin, Haemophilus influenzae; Lmo, Listeria monocytogenes; Msm, Mycobacterium smegmatis; Ngo, Neisseria gonorrhoeae; Nme, Neisseria meningitidis; Pae, Pseudomonas aeruginosa; Pfu, Pyrococcus furiosus; Rso, Ralstonia solanacearum; Sen, Salmonella enterica serovar Typhimurium; Sau, Staphylococcus aureus; Spn, Streptococcus pneumoniae; Vch, Vibrio cholerae; and Vpa, Vibrio parahaemolyticus. The scale bar represents 10% difference in amino acid sequences. Amino acid identities of each MATE protein as compared to FepA are indicated in square brackets. The two DinF and NorM subfamilies are highlighted.