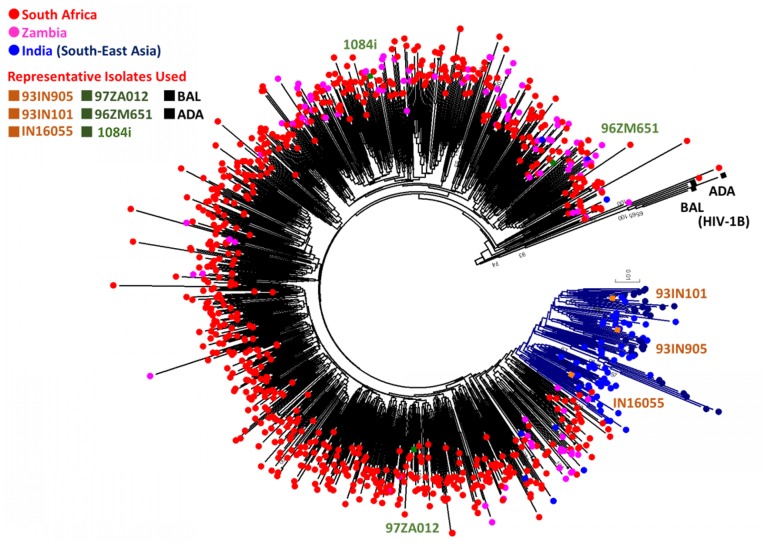
Figure 1. Phylogenetic analysis of gp120 sequences.
Neighbor-joining phylogenetic tree was constructed in MEGA 6.06 software using 878 HIV-1C and four HIV-1B (as outlier) gp120 sequences from India and from the Southern African Countries of South Africa and Zambia. Due to the comparatively fewer HIV-1C gp120 sequences that were available from India, we included HIV-1C gp120 sequences from China and Myanmar that were available in the database. A separate phylogenetic tree with HIV-1C gp120 sequences from China, Myanmar confirmed their Indian origin. HIV-1B sequences from US were included in the phylogenetic analysis for comparison. Bootstrap values >50% were shown in the tree. The HIV-1C gp120 sequences from India and Southern African countries are genetically distinct and cluster separately. The specific isolates used in our in-vitro analysis (HIV-1ADA, HIV-1Bal, HIV-1IndieC1, HIV-193IN905, HIV-1IN16055, HIV-11084i, HIV-197ZA012 and HIV-196ZM651) are included in this analysis (and indicated on the diagram) to show that they qualify to be used as representative isolates.