Figure 2. Bioinformatics analyses of siRNA screen hits.
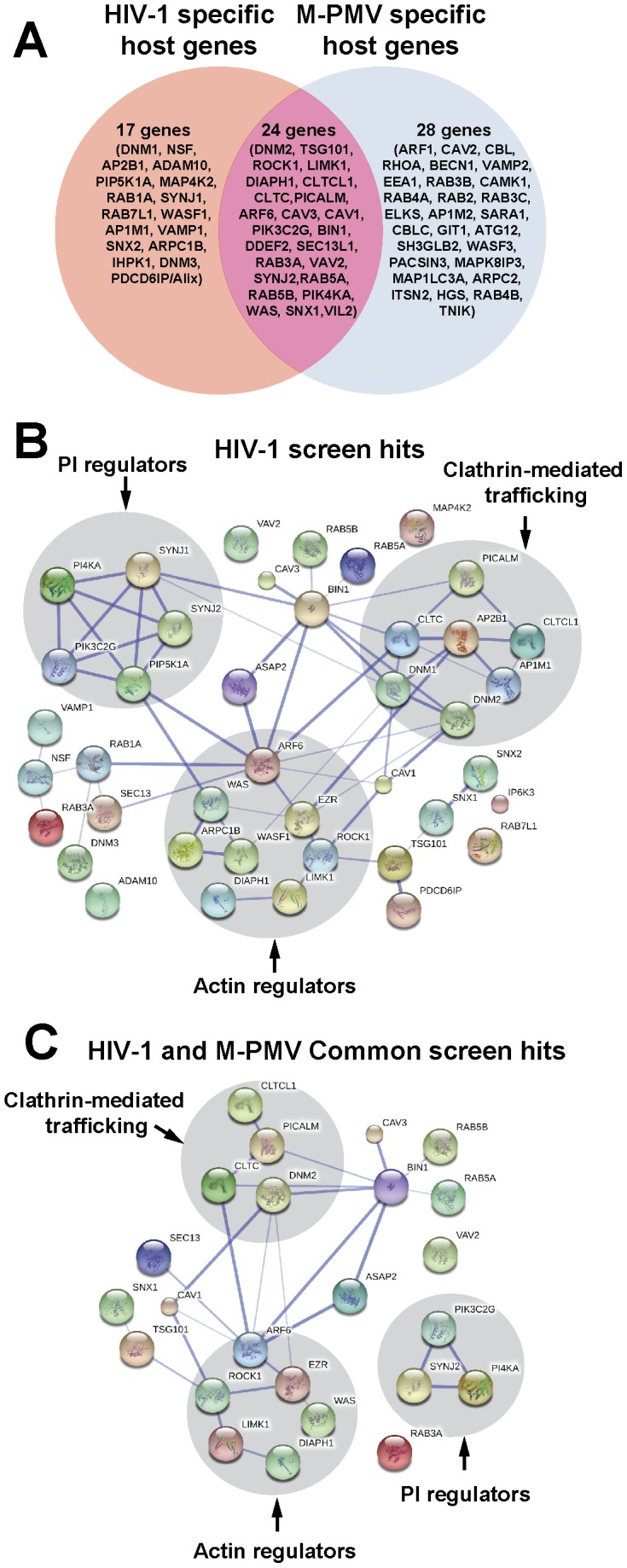
(A) Venn diagram of siRNA hits identified in this work. We identified 41 host proteins in HIV screen and 52 host proteins in M-PMV screen. There were 24 host proteins that were common for both HIV-1 and M-PMV. (B) Protein interaction network of HIV1 screen hits (Stronger associations are represented by thicker lines). A map of the interactions of our HIV screen hits was built by using STRING 9.05 with a required medium confidence score (0.400). Analysis of the network revealed clusters of interrelated factors involved in three distinct pathways (highlighted in grey circle): cellular actin cytoskeletal regulation pathways, clathrin-mediated trafficking factors and regulators of phosphoinositide metabolism. (C) Protein interaction network of the common screen hits for both HIV-1 and M-PMV (Stronger associations are represented by thicker lines). The interactions map of 24 host proteins that were common for both HIV-1 and M-PMV was built by using STRING 9.05 with a required medium confidence score (0.400). Factors involved in clathrin-mediated trafficking, regulation of the actin cytoskeleton, and phosphoinositide metabolism (highlighted in grey circle) were identified to affect both HIV-1 and M-PMV assembly or release.
