Figure 1.
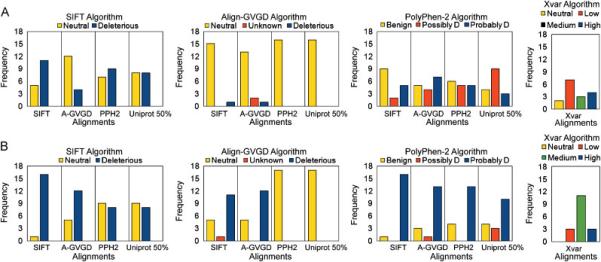
A) Predictions of neutral (n = 16) BRCA1 missense mutations using three algorithms with four alignments each. The four alignments are represented by SIFT (SIFT), Align-GVGD (A-GVGD), PolyPhen-2 (PPH2), and Uniprot 50% (Uniprot 50%). The prediction categories for PolyPhen-2 Possibly Damaging and Probably Damaging have been abbreviated to ‘Possibly D’ and ‘Probably D’. The algorithm Xvar employs its own alignment. B) Predictions of deleterious (n = 17) BRCA1 missense mutations using three algorithms with four alignments each.
