Abstract
Clear cell renal cell carcinoma (ccRCC) is the most common subtype of kidney cancer, and has the highest propensity to manifest as metastatic disease. Recent characterizations of the genetic signature of ccRCC have revealed several factors correlated with tumor cell migration and invasion; however the specific events driving malignancy are not well defined. Furthermore, there remains a lack of targeted therapies that result in long-term, sustainable response in patients with metastatic disease. We show here that neuronal pentraxin 2 (NPTX2) is over-expressed specifically in ccRCC primary tumors and metastases, and that it contributes to tumor cell viability and promotes cell migration through its interaction with the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunit GluR4. We propose NPTX2 as a novel molecular target for therapy for ccRCC patients diagnosed with or at risk of developing metastatic disease.
Keywords: neuronal pentraxin, clear cell renal cell carcinoma, invasion, metastasis, AMPA receptor, calcium
Introduction
Renal Cell Carcinoma (RCC) is one of the most common solid tumors in the United States, responsible for over 13,000 deaths annually (1). The clear cell variant (ccRCC) is the most common subtype of RCC, accounting for an estimated 80% of all patients (2). The prognosis for patients diagnosed with early stage disease is comparatively good, with stage I patients demonstrating over 90% and stage II patients demonstrating 63–95% overall survival of 5 or more years (3). Regrettably, up to 30% of early stage cases of ccRCC treated surgically will relapse with metastatic disease likely due to the presence of undetectable micrometastases (4). In addition, 20–30% of all ccRCC patients present with advanced or metastatic disease upon initial diagnosis oftentimes due to the asymptomatic nature of early stage disease (5).
Metastatic ccRCC renders a bleak prognosis, with an estimated 5 year overall survival of less than 10% due to lack of remedial therapies that produce significant disease regression or attenuation of disease progression (6). Drug resistance is a hallmark of ccRCC and is thought to be a culmination of several intrinsic and acquired tumorigenic properties linked to cancer cell heterogeneity, including a lack of known molecular factors which can be targeted pharmacologically (7, 8). ccRCC rarely responds to chemotherapy and radiation therapies, and drug resistance develops rapidly with application of targeted therapies (8). It is also apparent that ccRCC tumor cells demonstrate a disposition for increased migratory capacity, likely a major contributing factor to the development of tumor metastasis and disease relapse. A focus on identification of therapeutically targetable molecular factors for ccRCC is paramount.
Neuronal pentraxins belong to a class of secreted proteins characterized by their pentraxin protein domain. They are related to C-reactive protein (CRP), which is a serum protein that contributes to host defense and is expressed during acute phase inflammatory responses in mammals (9). NPTX2 is homologous to neuronal pentraxin 1 (NPTX1, NP1) and neuronal pentraxin receptor (NPTXR, NPR), both of which have been characterized in nervous system tissues (10). NPTX2 has a broader expression pattern, and is observed in nervous, testicular, pancreatic, skeletal muscle, heart, and hepatic tissues (11). NPTX1 and 2 form homomeric or heteromeric multimers with NPTXR which may function to bind the pentraxins to cell membrane surfaces (12). Previous research has identified a role for neuronal pentraxin molecules in neurite outgrowth and in synaptic plasticity of neuronal cells (13). This is thought to be mediated through clustering of the AMPA family of ionic glutamate receptors which form ion permeable channels during the development of excitatory synapses in neuronal cells (14–17) or by facilitating uptake of synaptic material during synapse remodeling (12, 18).
Here we demonstrate an essential tumor promoting role for NPTX2 in ccRCC. We establish NPTX2 as a stimulatory ligand that binds to the AMPA receptor subunit GluR4, which we find also to be overexpressed in ccRCC, leading to Ca2+ influx, actin cytoskeletal remodeling, and increased tumor cell migration. Inhibition of NPTX2 expression in ccRCC cells leads to decreased tumor cell proliferation, decreased tumor cell invasion, and induction of programmed cell death. Our results substantiate that NPTX2/GluR4 is a critical survival pathway for ccRCC and is a therapeutic candidate for patients with or at risk of developing metastatic ccRCC.
Materials and Methods
Cell Lines and Reagents
Human ccRCC cell lines: A498, Caki1, and Caki2 were purchased from ATCC. KIJ265T and RWV366T were established in the Copland laboratory as previously described (19). UMRC2 and UMRC6 cells were a kind gift from Dr. Bart Grossman (20). STR validation of all tumor cell lines was performed as previously described (19). Normal renal epithelial cells (NRE) include K347, K355, K359, RCJ58N, K366N, HRE152 (Copland Lab) and HK2 (ATCC). All cell lines were cultured in DMEM medium (Cellgro) containing 5% FBS (Hyclone) and 1x penicillin-streptomycin (Invitrogen) at 37° C in humidified conditions with 5% CO2. VHL mutational and deletional status were examined via DNA sequencing and multiplex ligation dependent probe amplification, respectively. Human recombinant NPTX2 (R-NPTX2) generated in HEK293 cells was purchased from OriGene. CFM-2 was purchased from Tocris Bioscience.
Lentivirus
MISSION shRNA pLKO.1 constructs (Sigma-Aldrich) were used to make self-inactivating shRNA lentiviruses for NPTX2 (NM_002523.1-1316s1c1, NM_002523.1-1623s1c1, NM_002523.1-804s1c1, NM_002523.1-855s1c1), GluR4 (NM_000829.1-1676s1c1, NM_000829.1-2145s1c1, NM_000829.1-2285s1c1, NM_000829.1-925s1c1), and a non-target (NT) random scrambled sequence control (SHC002). Lentivirus was prepared as previously described (19).
Transfections
The following plasmids were used: HA epitope-tagged human NPTX2 (pPM-hNPTX2-HA) (Applied Biosystems), and Flag epitope-tagged human GluR4 (Flag-GluR4-Flip-His-pcDNA3.1)-a generous gift from Dr. Kari Keinanen at the University of Helsinki, Helsinki, Finland (21).
RNA Isolation and QPCR
RNA extraction was performed as previously described (22). cDNA was prepared from purified RNA samples using the High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems) per manufacturer’s instruction. TaqMan® Fast Universal PCR Master Mix (Applied Biosystems) and TaqMan® FAM™ dye-labeled probes including: NPTX2[Hs00383983_m1], GRIA1[Hs00181348_m1], GRIA2[Hs00181331_m1], GRIA3[Hs01557466_m1], GRIA4[Hs00898778_m1], CDH1[Hs00170423_m1], SNAI1[Hs00195591_m1], SNAI2[Hs00161904_m1], CTNNB1[Hs00355049_m1], TGFB1[Hs00171257_m1], SPARC[Hs00234160_m1], VIM[Hs00185584_m1], and FN1[Hs00415006_m1] were combined with prepared cDNA samples to analyze relative mRNA expression via QPCR. POLR2A was used as a normalization control. Fold change values were compared between normal and tumor samples, NT scrambled and target lentiviral infected samples, and transfected vs. empty vector controls using the ΔΔCT method (23).
Gene Array Analysis, Pathway Analysis, and Database Meta-Analysis
Purified RNA samples were sent to the Mayo Clinic Advanced Genomic Technology Center Gene Expression Core where gene array expression analysis was performed using Affymetrix Human Genome U133 Plus 2.0 Array chip. Gene expression data (Gene Expression Omnibus Accession #GSE-53757) and the details of the data processing and methodology are previously described in (22). Pathway analysis was done using IPA (Ingenuity Systems). Meta-analysis was performed as previously described using NextBio data mining platform (24).
Western Blot Analysis
Cell protein extracts were prepared using RIPA lysis buffer containing 50 mM Tris, 5 mM EDTA, 150 mM NaCl, 0.1% SDS, 0.5% Deoxycholate, 1% NP40, protease inhibitor cocktail (Roche) and phosphatase inhibitor (Pierce). Tissue protein extracts were prepared from frozen samples using 1% SDS (Invitrogen) in 50mM pH 8.0 Tris buffer containing protease inhibitor cocktail (Roche) and phosphatase inhibitor (Pierce) with brief sonication on ice. Electrophoresis, transfer, blocking and antibody preparations were performed as previously described (19). Primary antibodies included: NPTX2 (Sigma-Aldrich- PRS4573) and β-actin (Sigma-Aldrich-A5441), GluR1 (Santa Cruz-sc-55509), GluR2 (Santa Cruz-sc-7601), p-CAMK1 (Santa Cruz-sc-373853), total CAMK1 (Abnova-PAB2769), GluR3(Cell Signaling-D47E3), PARP (Cell Signaling-9542), HA-tag (Cell Signaling-3724) and Flag-tag (Cell Signaling-8146), GluR4 (Millipore-AB1508), total AKT (Cell Signaling-2972), and p-AKT (R&D Systems-AF887). Secondary species-specific horseradish peroxidase-labeled antibodies (Jackson Immunoresearch) were applied, and Supersignal chemiluminescent kit (Pierce) was used to perform detection.
Immunoprecipitation Assay
Immunoprecipitation assay was performed using Protein G Mag Sepharose beads (GE Healthcare) according to manufacturer’s protocol using the cross-linking method. Antibodies include anti-HA (Cell Signaling-3724) and anti-Flag (Cell Signaling-8146). Cell protein extracts were prepared using 1% Triton X-100 (Sigma-Aldrich) in 50mM Tris pH 7.4 containing 150mM NaCl, 1mM EDTA (Promega), protease inhibitor cocktail (Roche) and phosphatase inhibitor (Pierce) on ice. Sample protein concentrations were diluted to1 µg/mL in lysis buffer. Gel Electrophoresis, membrane transfer, and antibody detection techniques applied are those as described in Protein Expression Analysis above.
Immunohistochemistry (IHC) and Immunofluorescence (IF)
Tissue microarrays (TMA) were prepared using formalin-fixed paraffin-embedded tissue samples procured from de-identified patients. This study was approved by the Mayo Institutional Review Board. All IHC scores are reported as H-scores. Imaging, staining, and analysis were performed as previously described (19) using the primary antibodies: NPTX2 (Sigma-Aldrich-PRS4573), NPTXR (R&D Systems-AF4414), and GluR4 (Millipore-AB1508). Cells were plated and fixed for IF as previously described (19). Primary antibodies included: NPTX2 (Pierce-PA512289), GluR4 (Millipore- AB1508), HA-tag (Cell Signaling-3724) and Flag-tag (Cell Signaling-8146), Fibronectin (BD Biosciences-610077). Fluorescently-labeled, species specific secondary antibodies (Sigma-Aldrich) were applied. 20–60x fluorescent images were obtained using an Olympus microscope (Olympus IX71).
Cell Death Analysis via Flow Cytometry
ccRCC cell lines were infected with NT vs. target NPTX2 or GluR4 shRNA lentiviral constructs overnight. Cells were selected using puromycin (Sigma) for 72 hours. 7 days post selection, both adhered and floating cells were collected with Accutase (Innovative Cell Technologies, Inc.), washed with DPBS, and suspended in 1x cold binding buffer (BD Pharmingen) at 1×106 cells/mL. Cells were stained with Propidium Iodide (BD Pharmingen), and cell death analysis was performed using an Accuri C6 flow cytometer (Accuri). Unstained NT control cells were used to set population parameters.
Calcium Green-1 AM Staining
ccRCC cells were plated at 1,000 cells/well in 96-well clear-bottom black plates (Corning, Inc.). Cells were washed 3x with DPBS, and Calcium Green-1 AM stain (Invitrogen) was applied at 5 µM in 5% CS-FBS DPBS for 45 minutes, protected from light. Cells were washed 3x with PBS and Ca2+ buffer containing135mM NaCl, 5mM KCl, 1mM each CaCl2 and MgCl2, 5.6mM glucose, 10mM Hepes, and 0.1% BSA at pH 7.4 (25) was added with/without specified concentrations of CFM-2 and/or R-NPTX2. Fmax: Ca2+ buffer with 2uM ionomycin (LC Labs) and 10 mM CaCl2. Fmin: 10mM EGTA and 10 mM MgCl2 in Ca2+ buffer without CaCl2 (26). A SpectraMax M5 (Molecular Devices), set at 531 nM emission and 506 nM excitation at 37°C, was used to measure fluorescence at specified time points. The following formula was used to determine the free calcium concentration in each population of cells: [Ca2+]free = Kd [(F-Fmin)/ (Fmax – F)] where the dissociation constant (Kd) for Calcium Green-1 AM is 190 nM (26)
Invasion Assay
Cells were starved overnight in 0.2% FBS DMEM. 5,000 cells were plated (in triplicate) with 0.25% BSA in the upper chamber and 5% FBS was the attractant in the lower chamber. BD Biocoat Matrigel Invasion Chambers (8µm pore) (BD Biosciences) were prepared per manufacturer’s protocol. After 20 hours, transwell inserts were fixed in 100% methanol and stained with 0.2% crystal violet/2% ethanol. Invasive cells were counted and 10x images were obtained using an Olympus microscope (Olympus IX71).
Statistics
Experimental values are presented as either percentage or fold change ± s.d. unless otherwise specified. Group comparisons (normal vs. tumor, NT vs. shRNA, control vs. treated, empty vector vs. expression vector) were analyzed using two-tailed paired Student's t-test with changes greater than 20% where p<0.05 being considered statistically significant.
Results
NPTX2 expression is tumor specific and is required for ccRCC viability
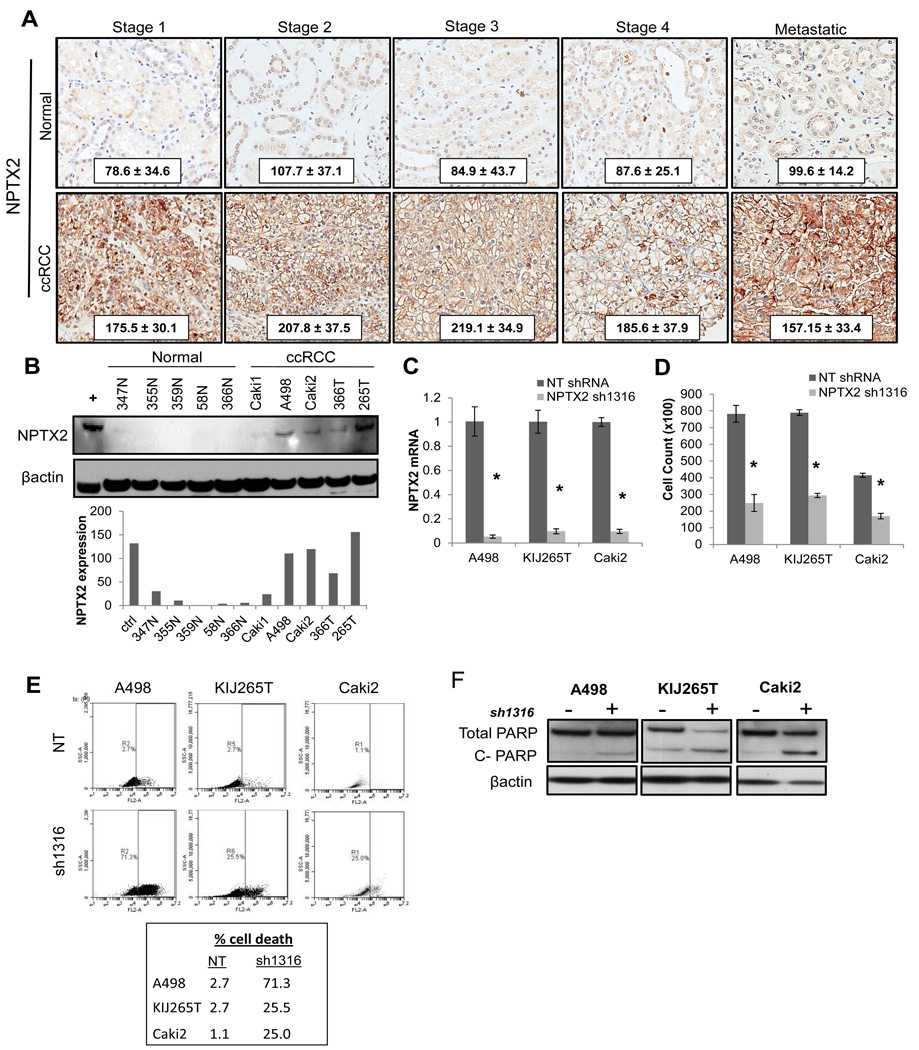
Patient gene array examining normal matched ccRCC patient mRNA from stage I, II, III, and IV revealed that NPTX2 expression was significantly increased in diseased samples at every stage (n=72 tumor and matched normal samples; GSE53757). To confirm these observations at the protein level, patient tissue microarrays were prepared from matched normal and ccRCC tissue samples from stage I, II, III, IV (primary tumor site), and tumor metastases. Immunohistochemistry (IHC) staining for NPTX2 confirmed significantly elevated protein levels in ccRCC samples at all stages as indicated by H scores (Fig 1A). Normal renal epithelial samples (NRE) and established ccRCC cell lines were examined for NPTX2 expression via QPCR and western blot analysis in order to establish working models. The majority of ccRCC cell lines demonstrate elevated NPTX2 at the mRNA level (5/7) when compared to normal samples (1/4) (SF 1A). Western blot analysis revealed high NPTX2 protein expression in A498, Caki2, and KIJ265T, lower NPTX2 expression in RWV366T, and little to no expression in Caki1 as well as all NRE cells examined (Fig 1B). The following cell lines are VHL mutant: Caki2, RWV366T, and KIJ265T while A498 and Caki1 are VHL wild type (SF 1B). The data suggest that NPTX2 expression in ccRCC is not related to von Hippel Lindau (VHL) gene status.
Figure 1. NPTX2 expression profile in clear cell Renal Cell Carcinoma.
(A) TMA IHC of patient ccRCC vs. matched normal tissue for NPTX2 expression in stage I, II, III, IV (primary), and IV (metastatic) (normal n=44, 32, 35, 7, and 6 and tumor n= 41, 26, 33, 10, and 17 respectively). Cytoplasmic and membranous staining pattern observed. H-score ± standard deviation from the mean is shown. (B) Western blot of protein lysates prepared from NRE cells and ccRCC cell lines for NPTX2 expression. Protein expression level quantitation is normalized to β-actin, and total human brain tissue lysate was used as a positive control. (C) NPTX2 knockdown was evaluated in A498, KIJ265T, and Caki2 ccRCC cell lines infected with the sh1316 lentiviral construct as compared to NT controls via QPCR. (D) 7 day proliferation assay of ccRCC sh1316 clones vs. NT controls. (E) Cell death of A498, KIJ265T, and Caki2 NT vs. sh1316 cell populations analyzed via flow cytometry. (F) Western blot for total and cleaved PARP in NT vs. NPTX2 knockdown cells. Cells collected for cell death analysis (panels E and F) correspond to day 7 of proliferation assay (panel D).
In order to examine the role of NPTX2 expression in ccRCC cell proliferation and viability, four lentiviral constructs designed to target NPTX2 were prepared (sh804, sh855, sh1316, and sh1623). A498 cells infected with each construct were evaluated by QPCR and western blot for lentiviral efficacy when compared to a non-target (NT) control (SF 1C,D). The NPTX2 shRNA-1316 (sh1316) construct demonstrated the highest knockdown, producing an 80% decrease in mRNA levels and resultant 60% decrease in protein levels (SF 1C,D). In order to validate lentiviral specificity for NPTX2, a rescue assay measuring proliferation was performed. A498 cells were transfected with an empty vector (EV) or a human NPTX2 expression plasmid (+ NPTX2), and were subsequently infected with either the NT or sh1316 lentiviral construct. sh1316 targets an intronic coding region of endogenous NPTX2 and thus does not affect recombinant NPTX2 plasmid overexpression in cells. NPTX2 overexpression was able to fully recover the proliferative capacity of A498 sh1316 cells (SF 1E).
A498, KIJ265T, and Caki2 cell lines, which express high endogenous NPTX2 (Fig 1B), were infected with the NPTX2 sh1316 construct. Over an 80% decrease in mRNA (Fig 1C) and 60% decrease in proliferation was observed 7 days post-selection in all three cell lines (Fig 1D). In order to evaluate the mechanism of decreased proliferation in ccRCC sh1316 cells, cell death was examined by flow cytometry. Results yielded a significant increase in cell death of A498, KIJ265T, and Caki2 sh1316 cell populations respectively when compared to NT controls (Fig 1E). Apoptosis was confirmed via western blot for PARP cleavage in NPTX2 knockdown cells (Fig 1F).
We used Nextbio (24) to assess publically available gene expression datasets (Table 1), and found that NPTX2 was consistently upregulated in nine comparisons of ccRCC vs. normal kidney tissue (27–35), and that NPTX2 was correlated specifically with the clear cell subtype of RCC as compared with other subtypes of RCC including granular, papillary, and chromophobe (Table 1)(36–38). We also found that NPTX2 was overexpressed in comparisons of more advanced ccRCC vs. less advanced disease including: stage IV vs. stage I, distant metastasis vs. no metastasis, metastasis vs. primary tumor, and stage II vs. stage I (Table 1)(35, 36, 38, 39). We performed a gene expression meta-analysis of the 13 top ccRCC experiments (27–30, 32, 33, 36, 37, 39–41) using Nextbio and found that NPTX2 was the most consistently differentially expressed gene (Table 2), being significantly up-regulated in all datasets. This data supports that NPTX2 overexpression is a prevalent feature of the ccRCC genetic profile, and is the most frequently dysregulated gene in patient tumor tissues.
Table 1. Expression analysis of NPTX2 in published microarray datasets.
NPTX2 expression was evaluated in nine datasets comparing normal versus ccRCC expression; datasets comparing expression of NPTX2 in the clear cell variant of RCC versus granular, papillary, and chromophobe RCC; and NPTX2 expression in various degrees of ccRCC disease progression (P<0.05).
| ccRCC vs. normal kidney | |||
|---|---|---|---|
| Difference | Significance | Reference | |
| Set 1 | 41.2 | 8.50E-05 | (27) |
| Set 2 | 33.6 | 1.10E-05 | (28) |
| Set 3 | 26.4 | 9.30E-05 | (29) |
| Set 4 | 39.2 | 0.0043 | (30) |
| Set 5 | 25.3 | 8.50E-12 | (31) |
| Set 6 | 11 | 1.60E-08 | (32) |
| Set 7 | 21 | 0.0002 | (33) |
| Set 8 | 9.35 | 0.037 | (34) |
| Set 9 | 104.8 | 0.0013 | (35) |
| Specificity for NPTX2 in ccRCC vs. other RCC subtypes | |||
| Difference | Significance | Reference | |
| ccRCC vs. granular RCC | 8.42 | 0.0007 | (36) |
| ccRCC vs. granular RCC | 31.8 | 2.00E-09 | (37) |
| ccRCC vs. papillary RCC | 12.3 | <1.0E-323 | (36) |
| ccRCC vs. papillary RCC | 30.6 | 3.90E-10 | (37) |
| ccRCC vs. papillary RCC | 9.09 | 6.00E-19 | (38) |
| ccRCC vs. chromophobe RCC | 4.64 | 8.00E-04 | (36) |
| Increased NPTX2 in more advanced ccRCC | |||
| Difference | Significance | Reference | |
| ccRCC stage IV vs. stage I | 6.98 | 8.00E-04 | (36) |
| ccRCC distant met vs. no met | 6.41 | 1.20E-03 | (36) |
| ccRCC met vs. primary tumor | 2 | 4.09E-02 | (39) |
| ccRCC stage II vs. stage I | 7.88 | 4.83E-01 | (35) |
| ccRCC stage II vs. stage I | 1.68 | 4.70E-03 | (38) |
Table 2. Meta-analysis of microarray data identifying commonly co-regulated genes in RCC.
The following comparisons were evaluated: A, ccRCC vs. normal; B, ccRCC vs. chromophobe RCC; C, ccRCC vs. granular RCC; D, ccRCC vs. papillary RCC. The top 10 most consistently differentially expressed genes in ccRCC are listed, and are significantly upregulated in all datasets evaluated (P<0.05).
| Comparison Type | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Score | A | A | A | A | A | A | A | A | B | B | B | C | D | |
| NPTX2 | 1219.94 | FC: | 26.4 | 11 | 41.2 | 39.2 | 21 | 35.8 | 33.6 | 35.9 | 16.7 | 4.64 | 8.42 | 12.3 | 2 |
| p-value: | 9.30E-05 | 1.60E-08 | 8.50E-05 | 0.0043 | 0.0002 | 3.30E-05 | 1.10E-05 | 0.0006 | 3.40E-09 | 0.0008 | 0.0007 | <1E-20 | 0.0409 | ||
| ANGPT2 | 1039.04 | FC: | 4.88 | 4.87 | 5.27 | 8.09 | 18.8 | 30.7 | 7.5 | 19.9 | 3.62 | 3.42 | 5.2 | 9.14 | 1.61 |
| p-value: | 2.20E-05 | 4.40E-15 | 0.0001 | 0.0374 | 0.0012 | 3.80E-07 | 5.10E-08 | 0.0117 | 1.70E-05 | 1.50E-05 | 0.0067 | <1E-20 | 0.035 | ||
| STC2 | 941.04 | FC: | 5.59 | 2.86 | 5.5 | 9.39 | 11.2 | 3.6 | 5.47 | 4.2 | 6.64 | 2.79 | 4.34 | 8.43 | 2.25 |
| p-value: | 0.0003 | 7.30E-09 | 1.70E-07 | 0.0305 | 1.50E-05 | 0.0002 | 2.90E-08 | 0.0001 | 0.0012 | 0.0005 | 0.0098 | 7.50E-42 | 0.0425 | ||
| CLEC2B | 768.065 | FC: | 3.94 | 3.2 | 3.38 | 5.6 | 3.06 | 5.31 | 3.27 | 3.82 | 2.82 | 2.93 | 4.71 | 2.48 | 1.66 |
| p-value: | 1.50E-07 | 7.00E-15 | 1.10E-05 | 0.0307 | 0.0072 | 7.80E-07 | 8.90E-09 | 0.0002 | 2.60E-05 | 0.001 | 0.027 | 4.70E-06 | 0.0126 | ||
| GBP1 | 755.053 | FC: | 2.63 | 3.09 | 3.1 | 2.54 | 2.65 | 4.24 | 3.6 | 3.59 | 7.35 | 3.63 | 10.7 | 3.43 | −1.44 |
| p-value: | 0.0012 | 6.70E-09 | 0.014 | 0.0082 | 0.0052 | 6.60E-05 | 0.0002 | 0.0016 | 9.60E-08 | 0.0026 | 0.0067 | 1.50E-08 | 0.0138 | ||
| PHKA2 | 710.102 | FC: | 2.56 | 2.71 | 3.37 | 5.47 | 3.27 | 4.09 | 3.24 | 5.64 | 4.98 | 1.91 | 2.72 | 3.15 | 1.72 |
| p-value: | 0.0003 | 3.20E-15 | 2.70E-11 | 0.0005 | 0.0071 | 8.10E-06 | 5.00E-10 | 0.0016 | 4.60E-06 | 0.0031 | 0.03 | 1.20E-17 | 0.0035 | ||
| PLXND1 | 618.956 | FC: | 1.95 | 2.1 | 1.92 | 2.29 | 8.87 | 3.37 | 2.36 | 4.4 | 5.05 | 2.54 | 2.72 | 3.03 | −1.4 |
| p-value: | 0.0004 | 4.30E-11 | 8.90E-05 | 0.0068 | 0.0028 | 3.10E-05 | 1.90E-08 | 0.0012 | 0.0103 | 3.80E-05 | 0.007 | 2.30E-12 | 0.0359 | ||
| CD300A | 586.535 | FC: | 2.78 | 2.14 | 2.62 | 2.61 | 2.94 | 4.38 | 2.4 | 15.8 | 7.85 | 1.65 | 2.8 | 1.32 | −1.42 |
| p-value: | 3.10E-11 | 2.00E-11 | 1.10E-05 | 0.0142 | 0.0038 | 5.60E-08 | 1.40E-05 | 0.009 | 0.0017 | 0.0043 | 0.0088 | 0.0329 | 0.0169 | ||
| HADH | 535.99 | FC: | −2.98 | −3.53 | −1.8 | −2.6 | −6.69 | −6.88 | −2.36 | −2.81 | −1.55 | −1.86 | −1.61 | −1.58 | −1.26 |
| p-value: | 0.0006 | 5.00E-15 | 0.0127 | 0.0341 | 7.90E-05 | 2.10E-06 | 2.60E-06 | 0.0335 | 0.0031 | 0.0002 | 0.0056 | 0.0005 | 0.0181 | ||
| CD99 | 513.844 | FC: | 2.03 | 2.39 | 1.77 | 4.09 | 1.84 | 2.83 | 2.06 | 2.01 | 2.19 | 2.64 | 4.94 | 1.56 | 1.45 |
| p-value: | 0.0265 | 7.50E-12 | 0.0017 | 0.0216 | 0.0093 | 0.0004 | 2.50E-05 | 0.0028 | 0.0057 | 9.00E-05 | 0.0337 | 8.60E-06 | 0.0325 | ||
| Reference: | (29) | (32) | (27) | (30) | (33) | (40) | (28) | (37) | (41) | (36) | (36) | (36) | (39) | ||
NPTX2 promotes invasive phenotype in ccRCC cells
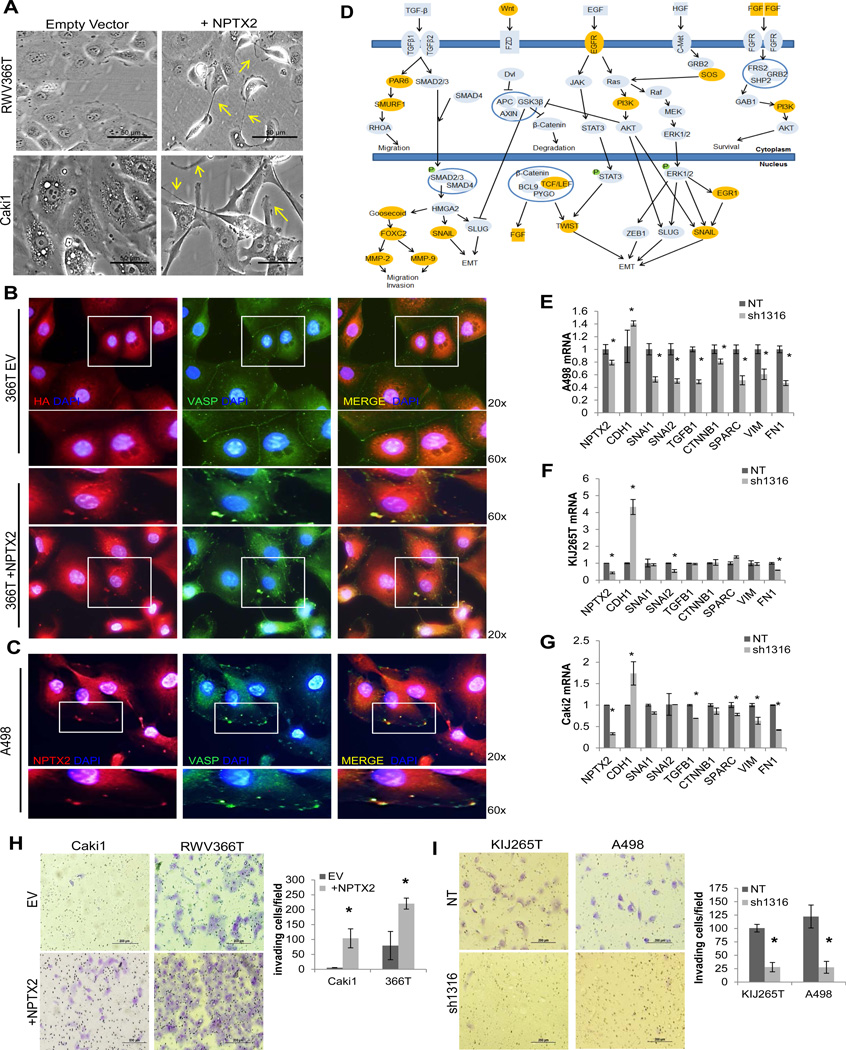
Immunofluorescence for NPTX2 demonstrates a specific protein expression pattern where it is most abundant at the leading cell edges and protrusions of the cell membrane as seen in KIJ265T cells which have high endogenous NPTX2 expression (SF 2A, Fig 1B). Over-expression of NPTX2 in RWV366T and Caki1 cells (+NPTX2) which have low endogenous NPTX2 (Fig 1B) induced morphological changes consistent with reduced cell-cell adhesion and increased cell migration, including the development of membrane protrusions as compared to empty vector (EV) control cells (Fig 2A). In order to evaluate whether NPTX2 expression is associated with actin cytoskeletal remodeling, immunofluorescence for VASP (vasodilator-stimulated phosphoprotein), an actin nucleation factor that localizes to areas of dynamic actin reorganization (42) was performed. In RWV366T EV control cells, VASP localizes along cell-cell boundaries (Fig 2B). In NPTX2 over-expressing RWV366T cells, VASP demonstrates a punctate expression pattern along the cell periphery (Fig 2B), where it co-localizes with ectopically expressed NPTX2. A498 ccRCC cells, which have high endogenous NPTX2 expression (Fig 1B), replicate the VASP staining pattern observed in 366T NPTX2-HA cells (Fig 2C). These results suggest that NPTX2 expression could alter actin dynamics and impair cell-cell adhesion in ccRCC cells.
Figure 2. NPTX2 modulates actin cytoskeletal remodeling and promotes invasion.
(A) Phase-contrast microscopy of empty vector vs. NPTX2 transfected (+NPTX2) RWV366T and Caki1 cells, taken at 20x magnification. Arrows are used to highlight select membranous protrusions. IF of (B) empty vector vs. NPTX2 (HA tagged) transfected (+NPTX2) RWV366T cells and (C) A498 cells stained for NPTX2 (left panel) and VASP (center panel). Images are merged in the right panel. 60x magnification of regions highlighted in 20x images are shown. (D) Pathway signature of EMT/cell migration in high-NPTX2 expressing tumors identified using Ingenuity Software analysis. Genes highlighted in orange are significantly upregulated in high-NPTX2 expressing tumors (P<0.05). QPCR for EMT-associated genes in NT vs. sh1316 NPTX2 knockdown (E) A498, (F) KIJ265T, and (G) Caki2 cells. Invasion assay of (H) empty vector vs. NPTX2 over-expressing Caki1 and RWV366T cells and (I) KIJ265T and A498 NT vs. sh1316 NPTX2 knockdown cells. Experiments were performed in triplicate, and representative images are displayed. Results are quantitated as invading cells per visual field.
Previous studies by our group have revealed that the genetic signature of ccRCC is associated with a loss of renal epithelial differentiation and increased expression of mesenchymal molecular markers (22), many of which have been previously implicated in tumor cell invasion (43). To examine the potential role of NPTX2 in ccRCC mesenchymal transformation and tumorigenesis, we initially performed pathway signature analysis. Using our ccRCC patient gene array (GSE53757), samples were sorted into high or low NPTX2 priori (SF 2B) and genes whose relative transcript expression was significantly altered (where fold change expression was ≤0.5 or ≥2) in the high NPTX2 group as compared to the low NPTX2 group were identified. Ingenuity® Software was used to perform pathway analysis. The results revealed NPTX2 co-regulation with a substantial number of genes associated with epithelial to mesenchymal transition (EMT) and cell migration (Fig 2D). To determine whether NPTX2 regulates the expression of these genes, QPCR for several EMT markers including CDH1, SNAI1, SNAI2, TGFB1, CTNNB1, SPARC, VIM, and FN1 (43) was performed in A498, KIJ265T, and Caki2 infected with either NT or sh1316 NPTX2 lentiviruses. All three cell lines demonstrated increased CDH1- a marker of epithelial differentiation (Fig 2E–G), and significant decreases in the expression of mesenchymal genes including: SNAI1, SNAI2, TGFB1, CTNNB1, SPARC, VIM, and FN1 in A498 cells (Fig 2E), SNAI2 and FN1 in KIJ265T cells (Fig 2F), and TGFB1, SPARC, VIM, and FN1 and Caki2 cells (Fig 2G) as a result of decreased NPTX2. Immunofluorescence for fibronectin (FN1), an EMT-associated extracellular matrix protein that was previously reported to contribute to tumor cell invasiveness (44) and that also correlates with an increased mortality rate in ccRCC patients (45), showed that A498 NT control cells exhibited high levels of both cytoplasmic and membranous fibronectin expression, and displayed numerous filopodia-like protrusions at the plasma membrane (SF 2C). Fibronectin expression at the cell periphery was significantly decreased in response to NPTX2 depletion, and the number of filopodia was greatly reduced (SF 2C). To test the effects of NPTX2 on invasive capabilities of ccRCC cells, invasion assays were performed. Caki1 EV cells exhibited little invasion, however when NPTX2 was overexpressed these cells demonstrated over a 10-fold increase in invasive potential (Fig 2H). Similarly, RWV366T NPTX2 cells displayed over a 2.5 fold increase in invasive potential as compared to EV control cells (Fig 2H). Conversely, KIJ265T and A498 sh1316 NPTX2 knockdown cells were considerably restricted in their ability to invade through the transwell inserts, with approximately a 70% decrease in KIJ265T and an 80% decrease in A498 sh1316 cells observed as compared to controls (Fig 2I). Of note, RWV366T and Caki1 control cells demonstrated lower overall invasion when compared to KIJ265T and A498 control cells (Fig 2H,I). Collectively, these results depict a correlation between NPTX2 expression and a migratory phenotype as well as implicate a role for NPTX2 in promoting ccRCC cell invasive capacity.
NPTX2 functions in ccRCC by binding to the AMPA receptor subunit GluR4
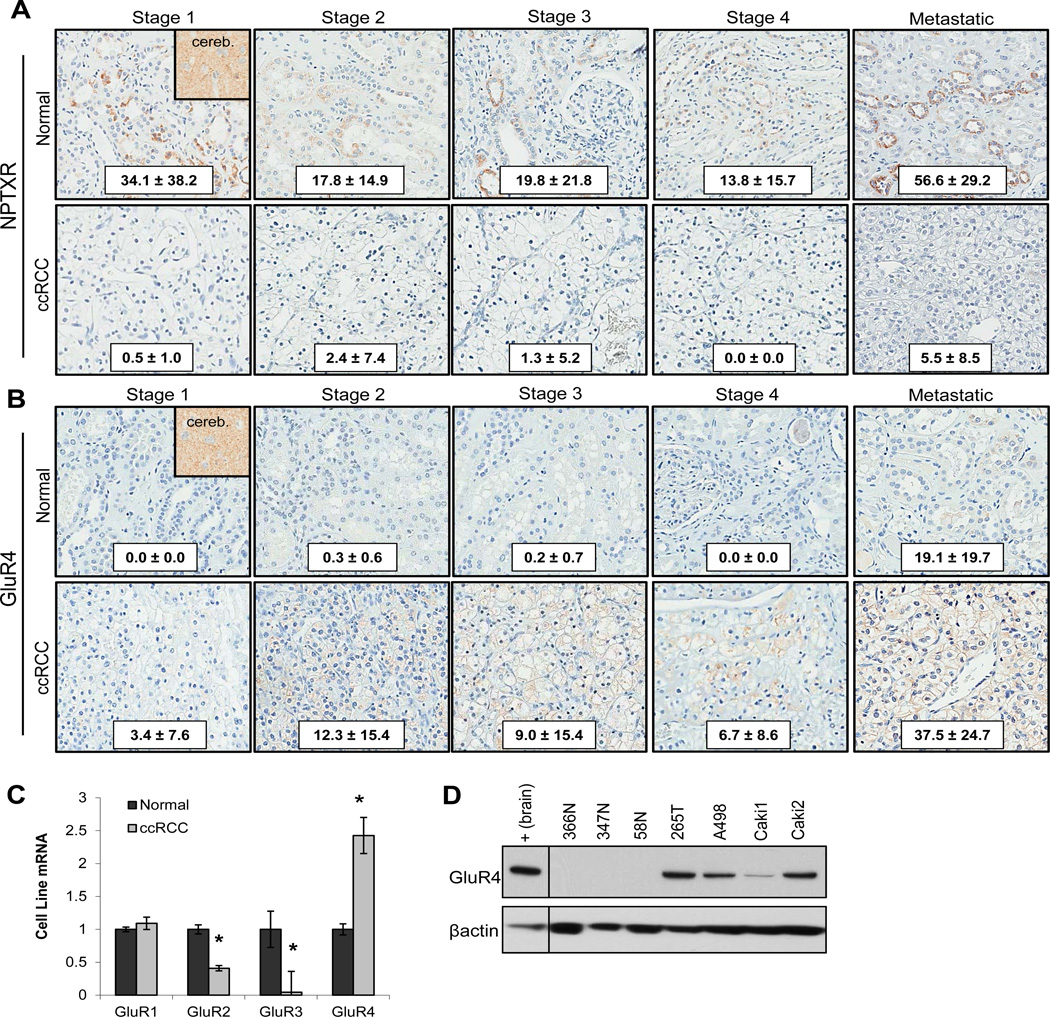
As NPTX2 is a secreted protein, we examined expression of the canonical receptor for NPTX2, neuronal pentraxin receptor (NPTXR) by IHC analysis, finding that ccRCC tissues demonstrate a loss of NPTXR expression when compared to matched normal kidney tissue, with very low detectable levels observed in ccRCC at any stage of disease (Fig 3A). We then evaluated expression of the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor subunits GluR1-4, as neuronal pentraxins have been previously reported to interact with this receptor complex, where it mediates clustering of AMPAR during synaptogenesis in excitatory neurons (15–17).
Figure 3. GluR4 is overexpressed in ccRCC.
(A) TMA IHC of patient ccRCC vs. matched normal tissue for NPTXR expression in stage I, II, III, IV (primary), and IV (metastatic) (normal n=12, 35, 34, 8, and 7 and tumor n= 13, 29, 34, 8, and 19 respectively). Human cerebellum tissue used as a positive control for NPTXR expression. (B) TMA IHC of patient ccRCC vs. matched normal tissue for GluR4 expression in stage I, II, III, IV (primary), and IV (metastatic) (normal n=45, 35, 38, 8, and 6 and tumor n= 39, 29, 34, 8, and 21 respectively). Cytoplasmic and membranous staining pattern observed. Human cerebellum tissue used as a positive control for GluR4 expression. H-score ± standard deviation from the mean is shown for NPTXR and GluR4 IHC analysis. (C) QPCR of NRE versus ccRCC cell lines for GluR1-4 (n=4 for both NRE and tumor samples). Tumor transcript expression is normalized to average NRE transcript expression. (D) Western blot of protein lysates prepared from NRE cells and ccRCC cell lines for GluR4 expression. Protein expression level quantitation is normalized to β-actin, and total human brain tissue lysate was used as a positive control.
Gene array revealed no change in GluR1 expression between normal and ccRCC, a decrease in GluR2 and GluR3 in ccRCC samples, and a 3-fold increase in GluR4 mRNA expression in ccRCC (GSE53757). Western blot of normal matched ccRCC metastatic tissue for GluR2 and 3 protein revealed no detectable levels in either matched normal or diseased tissue, and very low levels of GluR1 (Data Not Shown). GluR4 demonstrated a clear pattern of elevated tumor protein expression in 4 of 5 ccRCC samples via western blot (Data Not Shown). IHC of patient ccRCC tumor tissue stage I-IV and tumor metastasis revealed elevated GluR4 expression in ccRCC tissues as compared to normal kidney, with the highest expression observed in metastatic tissues (Fig 3B). In order to establish working cell models, NRE and ccRCC cells were evaluated by QPCR and WB for GluR4 expression. QPCR of normal and ccRCC cell lines demonstrated a similar pattern of AMPA receptor subunit expression observed in the patient gene array, with decreased expression of GluR2 and 3, no change in GluR1, and elevated GluR4 detected in tumor samples as compared to normal levels (Fig 3C). Three NRE samples and four ccRCC cell lines probed for GluR4 expression via western blot revealed no detectable protein expression in normal cells and high expression in tumor samples with high endogenous NPTX2 expression-KIJ265T, A498, and Caki2 (Fig 3D). Caki1 cells which have low endogenous NPTX2 also exhibit lower GluR4 expression (Fig 3D).
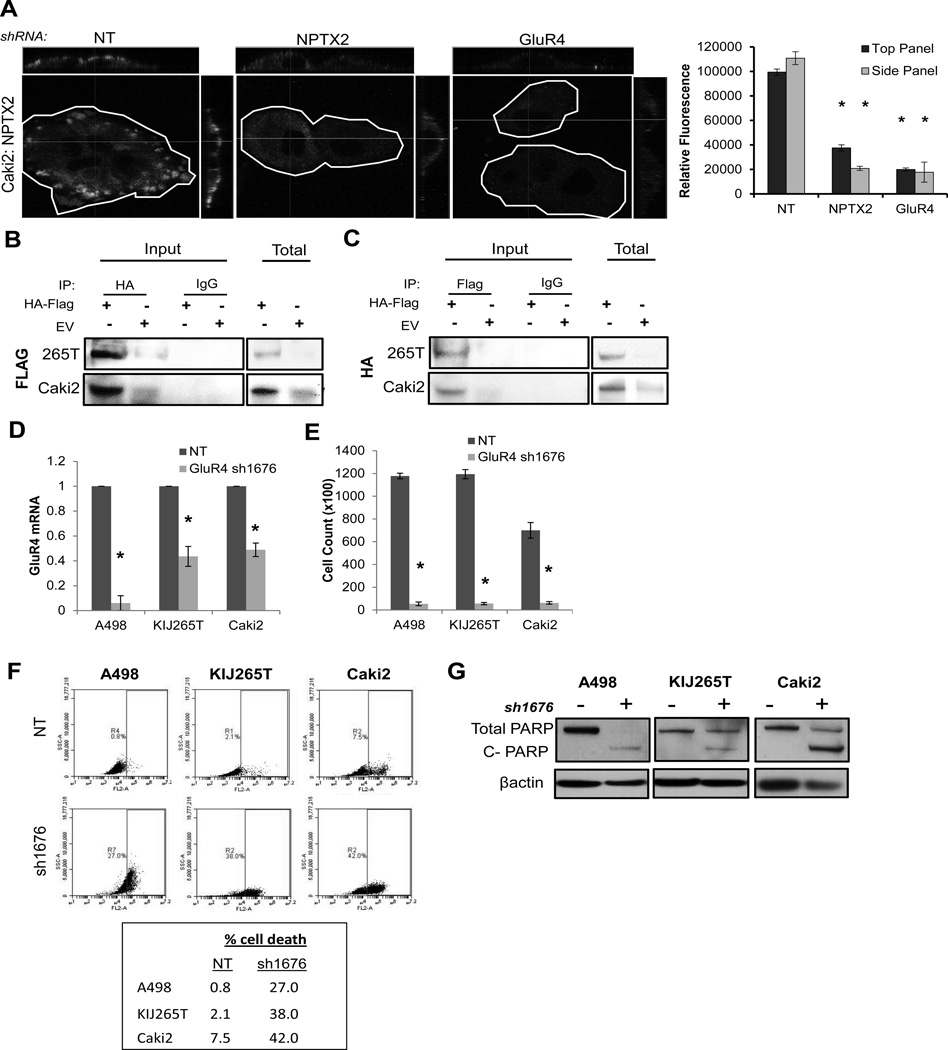
We next investigated whether GluR4 interacts directly with NPTX2 in ccRCC. Four lentiviral constructs targeting GluR4 were generated (sh925, sh1676, sh2145, sh2285), and efficacy was screened in A498 cells. The sh1676 construct yielded the most significant decrease in GluR4 mRNA and protein (SF 3A–B), and was therefore used in further experimentation. Caki2 cells, which express high levels of both NPTX2 and GluR4 (Fig 1B, Fig 3D) were examined for the ability of NPTX2 to adhere to the cell membrane in the presence or absence of GluR4. NT control cells exhibited a distinct pattern of NPTX2 clustering at the cell membrane (Fig 4A). In contrast, the sh1676 GluR4 knockdown cells demonstrated a similar pattern to that of sh1316 NPTX2 knockdown cells, with little to no NPTX2 binding observed at the cell membrane (Fig 4A). These results suggest that GluR4 is necessary for NPTX2 membrane adhesion in ccRCC cells. We also evaluated the interaction of NPTX2 and GluR4 via immunoprecipitation (IP). KIJ265T and Caki2 cells were transfected with epitope tagged human NPTX2-HA and GluR4-Flag expression vectors (HA-Flag). An IP was performed using an HA-tag specific antibody, and the resulting pull-downs were probed for GluR4 via western blot using a Flag-tag specific antibody. NPTX2 successfully bound GluR4 in both cell lines (Fig 4B). The reciprocal was also performed: pull-downs using a Flag-tag specific antibody followed by evaluation of associated NPTX2 expression using an HA-tag specific antibody demonstrated that GluR4 was able to co-precipitate NPTX2 in both cell lines (Fig 4C). These results confirm the interaction of NPTX2 and GluR4 in ccRCC.
Figure 4. GluR4 binds NPTX2 and promotes ccRCC cell viability.
(A) Confocal immunofluorescence of non-permeabilized Caki2 NT, sh1316, and sh1676 clones for NPTX2 binding to the cell membrane. Each cell is outlined, and cross sections are displayed in the upper and right panels for each image. Relative fluorescence corresponding to NPTX2 expression is quantitated. IP of KIJ265T and Caki2 cells transfected with both HA epitope tagged human NPTX2 and Flag epitope tagged human GluR4 (HA-Flag) versus empty vector (EV) for (B) HA (NPTX2) mediated Flag (GluR4) pulldown and (C) Flag (GluR4) mediated HA (NPTX2) pulldown. WB of total lysate (Total) confirms expression of epitope tagged proteins. (D) QPCR for GluR4 expression in A498, KIJ265T, and Caki2 NT vs. sh1676 GluR4 knockdown cell populations. (E) 7 day proliferation assay of A498, KIJ265T, and Caki2 NT vs. sh1676 clones. (F) Cell death of A498, KIJ265T, and Caki2 NT vs. sh1676 cell populations analyzed via flow cytometry. (G) Western blot for total and cleaved PARP in A498, KIJ265T, and Caki2 NT vs. GluR4 knockdown cells. Cells collected for cell death analysis (panels F and G) correspond to day 7 of proliferation assay (panel E).
Using the sh1676 lentiviral construct, GluR4 expression was knocked down in A498, KIJ265T, and Caki2 cells (Fig 4D), and resulting effects on proliferation and cell viability were evaluated. A significant reduction in proliferation (Fig 4E) and viability (Fig 4F, G) was observed as a result of GluR4 knockdown in all three ccRCC cell lines, demonstrating that loss of GluR4 in ccRCC phenocopied loss of NPTX2.
NPTX2-GluR4 mediates influx of intracellular calcium in ccRCC cells
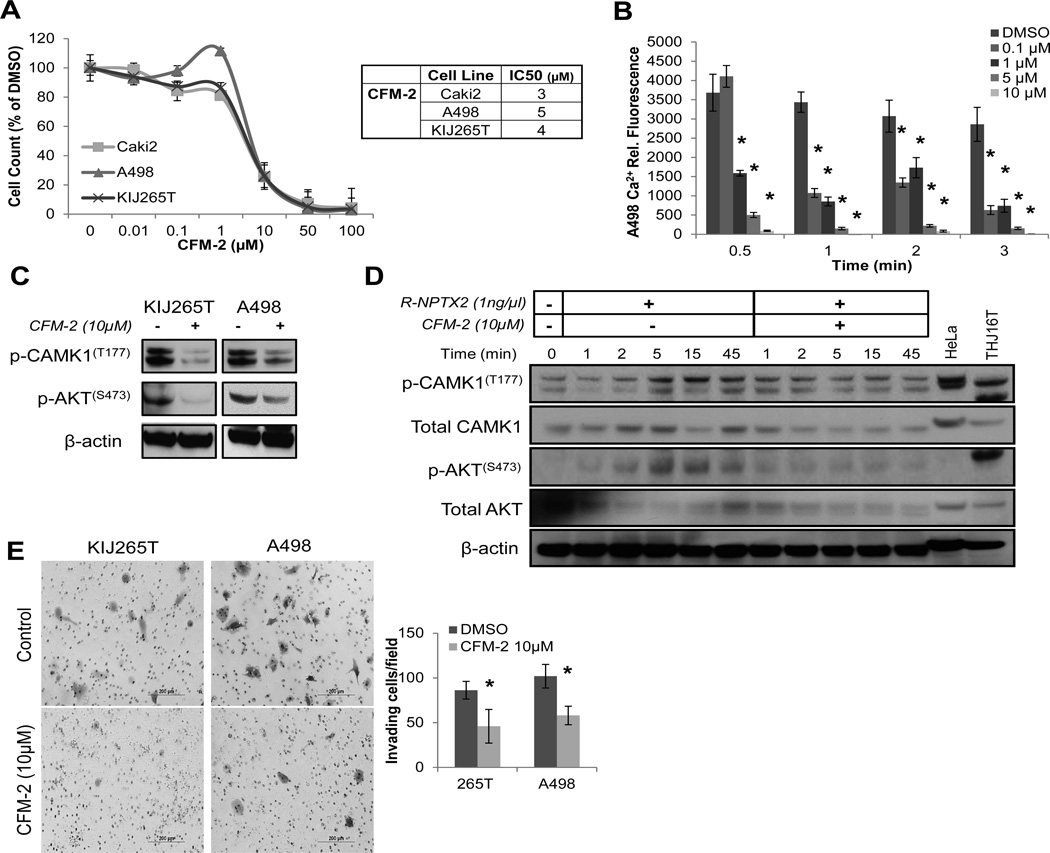
AMPA receptors belong to the non-NMDA-type of ionotropic-glutamate (iGluR) transmembrane receptors, which form homomeric or heterotrimeric calcium permeable ligand-gated ion channels composed of four subunits (46, 47). In order to evaluate whether NPTX2 plays a role in AMPA receptor mediated intracellular calcium influx, the allosteric AMPA antagonist CFM-2 (48), was tested in ccRCC cell lines to assess its effect on cell proliferation. Exposure to CFM-2 caused a dose-dependent anti-proliferative effect in ccRCC cell lines, with IC50 values in the low µM range (3–5µM) for Caki2, A498, and KIJ265T (Fig 5A).
Figure 5. NPTX2-GluR4 mediates influx of intracellular calcium in ccRCC cells.
(A) Proliferation of ccRCC cells in response to CFM-2 treatment (AMPA receptor antagonist) at day 7. (B) Effects of CFM-2 on A498 intracellular calcium levels using the cell permeable fluorescently labeled calcium indicator Calcium Green™-1, AM. Results are calculated as change in fluorescence over time (min). (*) Denotes significant change in fluorescence as compared to DMSO control per time point. (C) Western blot of KIJ265T and A498 cells treated with DMSO vs. CFM-2 (10µM, 1 hour) for phosphorylation of CAMK1(T177) and AKT(S473). (D) Western blot of R-NPTX2 (1ng/µL) treated Caki1 cells for phosphorylation of CAMK1(T177) and AKT(S473) treated with DMSO vs. CFM-2 (10µM) over time (min). (E) Invasion assay of DMSO vs. CFM-2 (10µM) treated KIJ265T and A498 cells. Experiments were performed in triplicate, and representative images are displayed. Results are quantitated as invading cells per visual field.
In order to evaluate the effect of NPTX2 on ccRCC intracellular calcium levels, A498 cells, which have high endogenous NPTX2 and GluR4 (Fig 1B, 3D), were stained with Calcium Green™-1, treated with CFM-2, and fluorescence relative to intracellular Ca2+ content was measured. CFM-2 induced a dose-dependent decrease in intracellular Ca2+ in A498 cells (Fig 5B). We also examined the effects of exogenous application of recombinant human NPTX2 protein (R-NPTX2) on Caki1 intracellular Ca2+ influx. Treatment of empty vector transfected Caki1 cells with R-NPTX2 (1ng/µL) increased intracellular Ca2+ influx by approximately 6-fold at 1 minute, and this was partially blocked by CFM-2 (10µM) (SF 4A). These effects were greatly enhanced in GluR4 transfected Caki1 cells (SF 4B). Similarly, co-transfection of NPTX2 and GluR4 into Caki1 cells further demonstrate sustained elevated Ca2+ as compared to the EV control or NPTX2 alone (SF 4C). CFM-2 treatment resulted in a dose-dependent decrease of intracellular Ca2+ in both NPTX2 and NPTX2-GluR4 transfected Caki1 cells, with the most significant reductions observed at 10µM (SF 4C). The phosphorylation of CAMK1 and AKT kinases and subsequent activation are modulated by intracellular calcium, and both kinases are involved in AMPAR signaling and regulation (49, 50). Phosphorylation of CAMK1 at Thr177(51) and AKT at S473(52) are reported to represent full activation of each kinases. KIJ265T and A498 cells were treated with a 10µM dose of CFM-2 for 1 hour, and CAMK1 and AKT phosphorylation was evaluated via western blot. CAMK1 and AKT phosphorylation was significantly reduced in response to CFM-2 treatment in both cell lines (Fig 5C). Caki1 cells treated with 1ng/µL of R-NPTX2 were probed for phosphorylation of CAMK1 and AKT over a time course. Increases in CAMK1 and AKT phosphorylation were observed at 1 minute and peaked at 5 minutes post R-NPTX2 treatment (Fig 5D). R-NPTX2 mediated downstream phosphorylation was completely blocked by CFM-2 treatment (Fig 5D). These data demonstrate a role for NPTX2 in mediating activation of downstream calcium-dependent kinases such as CAMK1 and AKT, likely by facilitating calcium influx through AMPA receptor complexes.
As our data suggests a role for NPTX2 in ccRCC cell invasiveness, we also tested the effects of CFM-2 treatment on KIJ265T and A498 in vitro invasion. Cells were plated in invasion chambers with or without CFM-2 treatment (10µM), and invading cells were stained and counted after 20 hours. CFM-2 treatment reduced both KIJ265T and A498 cell invasive capacity by approximately 50% (Fig 5E). These data show that AMPA receptor activity influences ccRCC cell invasion, and that this may be abrogated with treatment using an AMPAR antagonist such as CFM-2.
Discussion
Here we identify overexpression of NPTX2 as a novel feature of ccRCC, with elevated transcript and protein expression detected at all stages of disease in the majority of samples examined (Fig 1). Meta-analysis of public datasets revealed that NPTX2 is consistently up-regulated at the transcript level in ccRCC, and that NPTX2 expression is more strongly correlated with advanced ccRCC lesions versus low grade lesions (Table 1). Interestingly, of the other top 10 co-regulated genes significantly altered in ccRCC (Table 2), each of them have been associated with roles in cell migration, inflammation, angiogenesis, and metabolism- pathways implicated in oncogenesis. Several have been identified as pro-tumorigenic including ANGPT2 (53), STC2 (54), CLEC2B (55), GBP1 (56), and PLXND1 (57). Previous studies in pancreatic cancer have implicated NPTX2 as having tumor suppressor activity (58). We show here that in the context of ccRCC NPTX2 supports tumor cell viability, correlates with a pro-migratory phenotype, and additionally promotes ccRCC cell invasion in an in vitro setting (Fig 2).
We further identify GluR4 as a receptor for NPTX2 (Fig 3, 4) in ccRCC. In a normal physiological context GluR4 expression appears to be predominantly localized to specific neuronal and glial cells in the central nervous system (CNS) where it mediates fast synaptic neurotransmission (17, 59, 60). Current investigations in the role of AMPA receptors have additionally defined this group of receptors as fundamental for neuronal cell migration in the developing CNS (61, 62). ccRCC is thought to arise from the epithelial cells of the proximal renal tubule (63), for which there currently is no defined role for AMPA receptors in a normal context of this cell type. Therefore, the AMPA receptor expression profile in ccRCC is unique, and our findings show that GluR4 is important for ccRCC viability and invasion. Recently, AMPA receptor activity has been linked to neuronal cancer cell migration (64–66). Furthermore, glutamate, the excitatory neurotransmitter for two major receptor families: the metabotropic-glutamate (mGluR) and ionotropic-glutamate (iGluR) receptors, is linked to cell proliferation and metastasis in a number of cancers (62, 67–69). Taken together, our data lend support to an important pro-survival role of AMPA receptors in ccRCC. The regulation of this receptor complex warrants further exploration to define its role in the development and/or progression of ccRCC.
NPTX2 induces intracellular calcium flow in ccRCC cells in a GluR4 dependent manner which can be suppressed via treatment with CFM-2, an allosteric AMPA receptor antagonist. Furthermore, CFM-2 treatment reduces invasion in ccRCC cells with high endogenous NPTX2 expression (Fig 5). These results suggest a role for NPTX2 in promoting ccRCC tumor cell migration, in part facilitated downstream through its interaction with GluR4. Recent literature establishes that intracellular calcium dynamics contribute to cell mobilization. Localized Ca2+ gradients, or calcium flickers, can be found at the leading edges of migrating fibroblasts, where they are thought to regulate calcium-dependent actin cytoskeletal re-organization (70, 71). We propose that NPTX2 mediated influx of Ca2+ via GluR4 expressing AMPAR complexes results in reorganization of the actin cytoskeleton, and promotes cell migration. In addition to reduced tumor cell migration, decreased NPTX2 expression leads to attenuated tumor cell proliferation and induction of cell death of ccRCC cells. Similar results are observed with decreased GluR4 in ccRCC cells. While our results demonstrate a role for NPTX2 in ccRCC tumor cell viability and migration, further characterization of its interaction with AMPAR as well as delineation of immediate downstream signaling needs to be investigated.
Collectively, our results demonstrate that NPTX2 is a novel tumor-specific factor which demonstrates a consistent pattern of overexpression in the clear cell subtype of RCC, whose activity mediates tumor cell survival and invasion. We propose NPTX2 as a strong candidate for targeted therapy, whose inhibition may demonstrate a clinical benefit in patients suffering from metastatic ccRCC. Additionally, NPTX2 may serve as a predictive biomarker in patients presenting with localized disease at risk of developing metastatic disease, and subsequently should also be investigated as a preventative therapeutic target. As NPTX2 is a secreted protein, development of neutralizing antibodies may provide a feasible therapeutic option. Furthermore, we identify GluR4 as a downstream mediator of NPTX2 activity whose expression also appears to be vital for ccRCC viability, and additionally should be investigated as a therapeutic target.
Supplementary Material
Acknowledgements
We would like to thank Brandy Edenfield for processing and staining immunohistochemistry tissues. We also recognize James L. Miller, Michele M. Hoffmann, and Laura J. Lewis-Tuffin for their technical support.
Funding support:
This work was funded in part from NIH/NCI grants R01CA104505, R01CA136665, R01CA104505-05S1 (JAC); a generous gift from the David & Lois Stulberg Endowed Fund for Kidney Cancer Research (JAC); Mr. and Mrs. Ompal Chauhan Research Fund (JAC); Kidney Cancer Research at Mayo Clinic in Florida (JAC); James C. and Sarah K. Kennedy Mayo Clinic Research Career Development Award for Clinicians (HWT); Scheidel Foundation (JAC); Fraternal Order of Eagles Florida State Auxiliary (JAC); and a grant for rare cancers from Dr. Ellis and Dona Brunton (JAC).
Footnotes
Conflicts of interest:
The authors of this manuscript have no conflicts of interest to disclose.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, et al. Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J Clin Oncol. 2005;23:2763–2771. doi: 10.1200/JCO.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 3.Lam JS, Shvarts O, Pantuck AJ. Changing concepts in the surgical management of renal cell carcinoma. Eur Urol. 2004;45:692–705. doi: 10.1016/j.eururo.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Passebosc-Faure K, Lambert C, Gentil-Perret A, Blanc F, Oosterwijk E, et al. The expression of G250/mn/CA9 antigen by flow cytometry: its possible implication for detection of micrometastatic renal cancer cells. Clin Cancer Res. 2001;7:89–92. [PubMed] [Google Scholar]
- 5.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 6.Haddad H, Rini BI. Current treatment considerations in metastatic renal cell carcinoma. Curr Treat Options Oncol. 2012;13:212–229. doi: 10.1007/s11864-012-0182-8. [DOI] [PubMed] [Google Scholar]
- 7.Motzer RJ, Bacik J, Mazumdar M. Prognostic factors for survival of patients with stage IV renal cell carcinoma: memorial sloan-kettering cancer center experience. Clin Cancer Res. 2004;10:6302S–6303S. doi: 10.1158/1078-0432.CCR-040031. [DOI] [PubMed] [Google Scholar]
- 8.Coppin C, Kollmannsberger C, Le L, Porzsolt F, Wilt TJ. Targeted therapy for advanced renal cell cancer (RCC): a Cochrane systematic review of published randomised trials. BJU Int. 2011;108:1556–1563. doi: 10.1111/j.1464-410X.2011.10629.x. [DOI] [PubMed] [Google Scholar]
- 9.Romero IR, Morris C, Rodriguez M, Du Clos TW, Mold C. Inflammatory potential of C-reactive protein complexes compared to immune complexes. Clin Immunol Immunopathol. 1998;87:155–162. doi: 10.1006/clin.1997.4516. [DOI] [PubMed] [Google Scholar]
- 10.Omeis IA, Hsu YC, Perin MS. Mouse and human neuronal pentraxin 1 (NPTX1): conservation, genomic structure, and chromosomal localization. Genomics. 1996;36:543–545. doi: 10.1006/geno.1996.0503. [DOI] [PubMed] [Google Scholar]
- 11.Hsu YC, Perin MS. Human neuronal pentraxin II (NPTX2): conservation, genomic structure, and chromosomal localization. Genomics. 1995;28:220–227. doi: 10.1006/geno.1995.1134. [DOI] [PubMed] [Google Scholar]
- 12.Kirkpatrick LL, Matzuk MM, Dodds DC, Perin MS. Biochemical interactions of the neuronal pentraxins. Neuronal pentraxin (NP) receptor binds to taipoxin and taipoxin-associated calcium-binding protein 49 via NP1 and NP2. J Biol Chem. 2000;275:17786–17792. doi: 10.1074/jbc.M002254200. [DOI] [PubMed] [Google Scholar]
- 13.Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes C, Worley PF. Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci. 1996;16:2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, et al. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 16.O'Brien R, Xu D, Mi R, Tang X, Hopf C, Worley P. Synaptically targeted narp plays an essential role in the aggregation of AMPA receptors at excitatory synapses in cultured spinal neurons. J Neurosci. 2002;22:4487–4498. doi: 10.1523/JNEUROSCI.22-11-04487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 18.Dodds DC, Omeis IA, Cushman SJ, Helms JA, Perin MS. Neuronal pentraxin receptor, a novel putative integral membrane pentraxin that interacts with neuronal pentraxin 1 and 2 and taipoxin-associated calcium-binding protein 49. J Biol Chem. 1997;272:21488–21494. doi: 10.1074/jbc.272.34.21488. [DOI] [PubMed] [Google Scholar]
- 19.von Roemeling CA, Marlow LA, Wei JJ, Cooper SJ, Caulfield TR, Wu K, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19:2368–2380. doi: 10.1158/1078-0432.CCR-12-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossman HB, Wedemeyer G, Ren LQ. Human renal carcinoma: characterization of five new cell lines. J Surg Oncol. 1985;28:237–244. doi: 10.1002/jso.2930280320. [DOI] [PubMed] [Google Scholar]
- 21.Coleman SK, Cai C, Mottershead DG, Haapalahti JP, Keinanen K. Surface expression of GluR-D AMPA receptor is dependent on an interaction between its C-terminal domain and a 4.1 protein. J Neurosci. 2003;23:798–806. doi: 10.1523/JNEUROSCI.23-03-00798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5:e10696. doi: 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 24.Kupershmidt I, Su QJ, Grewal A, Sundaresh S, Halperin I, Flynn J, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS One. 2010;(5) doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz C, Perraud AL. The TRPM cation channels in the immune context. Curr Pharm Des. 2005;11:2765–2778. doi: 10.2174/1381612054546851. [DOI] [PubMed] [Google Scholar]
- 26.Silei V, Fabrizi C, Venturini G, Tagliavini F, Salmona M, Bugiani O, et al. Measurement of intracellular calcium levels by the fluorescent Ca(2+) indicator Calcium-Green. Brain Res Brain Res Protoc. 2000;5:132–134. doi: 10.1016/s1385-299x(00)00003-9. [DOI] [PubMed] [Google Scholar]
- 27.Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 28.Kort EJ, Farber L, Tretiakova M, Petillo D, Furge KA, Yang XJ, et al. The E2F3-Oncomir-1 axis is activated in Wilms' tumor. Cancer Res. 2008;68:4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Roche O, Yan MS, Finak G, Evans AJ, Metcalf JL, et al. Regulation of endocytosis via the oxygen-sensing pathway. Nat Med. 2009;15:319–324. doi: 10.1038/nm.1922. [DOI] [PubMed] [Google Scholar]
- 30.Stickel JS, Weinzierl AO, Hillen N, Drews O, Schuler MM, Hennenlotter J, et al. HLA ligand profiles of primary renal cell carcinoma maintained in metastases. Cancer Immunol Immunother. 2009;58:1407–1417. doi: 10.1007/s00262-008-0655-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusenko MV, Zubakov D, Kovacs G. Gene expression profiling of chromophobe renal cell carcinomas and renal oncocytomas by Affymetrix GeneChip using pooled and individual tumours. Int J Biol Sci. 2009;5:517–527. doi: 10.7150/ijbs.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 33.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 35.Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A, Leng N, Pavia-Jimenez A, Wang S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.(ICG) TIGC. The expO project (Expression Project for Oncology) [cited; Available from: www.intgen.org.
- 37.Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beleut M, Zimmermann P, Baudis M, Bruni N, Buhlmann P, Laule O, et al. Integrative genome-wide expression profiling identifies three distinct molecular subgroups of renal cell carcinoma with different patient outcome. BMC Cancer. 2012;12:310. doi: 10.1186/1471-2407-12-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan X, Zhai Y, Chang W, Hou J, He S, Lin L, et al. Global analysis of metastasis-associated gene expression in primary cultures from clinical specimens of clear-cell renal-cell carcinoma. Int J Cancer. 2008;123:1080–1088. doi: 10.1002/ijc.23637. [DOI] [PubMed] [Google Scholar]
- 40.Cifola I, Spinelli R, Beltrame L, Peano C, Fasoli E, Ferrero S, et al. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol Cancer. 2008;7:6. doi: 10.1186/1476-4598-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams AA, Higgins JP, Zhao H, Ljunberg B, Brooks JD. CD 9 and vimentin distinguish clear cell from chromophobe renal cell carcinoma. BMC Clin Pathol. 2009;9:9. doi: 10.1186/1472-6890-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause M, Dent EW, Bear JE, Loureiro JJ, Gertler FB. Ena/VASP proteins: regulators of the actin cytoskeleton and cell migration. Annu Rev Cell Dev Biol. 2003;19:541–564. doi: 10.1146/annurev.cellbio.19.050103.103356. [DOI] [PubMed] [Google Scholar]
- 43.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen SH, Lin CY, Lee LT, Chang GD, Lee PP, Hung CC, et al. Up-regulation of fibronectin and tissue transglutaminase promotes cell invasion involving increased association with integrin and MMP expression in A431 cells. Anticancer Res. 2010;30:4177–4186. [PubMed] [Google Scholar]
- 45.Steffens S, Schrader AJ, Vetter G, Eggers H, Blasig H, Becker J, et al. Fibronectin 1 protein expression in clear cell renal cell carcinoma. Oncol Lett. 2012;3:787–790. doi: 10.3892/ol.2012.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32:841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 47.Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chimirri A, De Sarro G, De Sarro A, Gitto R, Grasso S, Quartarone S, et al. 1-Aryl-3,5-dihydro-4H-2,3-benzodiazepin-4-ones: novel AMPA receptor antagonists. J Med Chem. 1997;40:1258–1269. doi: 10.1021/jm960506l. [DOI] [PubMed] [Google Scholar]
- 49.Ishiuchi S, Yoshida Y, Sugawara K, Aihara M, Ohtani T, Watanabe T, et al. Ca2+-permeable AMPA receptors regulate growth of human glioblastoma via Akt activation. J Neurosci. 2007;27:7987–8001. doi: 10.1523/JNEUROSCI.2180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fortin DA, Davare MA, Srivastava T, Brady JD, Nygaard S, Derkach VA, et al. Long-term potentiation-dependent spine enlargement requires synaptic Ca2+-permeable AMPA receptors recruited by CaM-kinase I. J Neurosci. 2010;30:11565–11575. doi: 10.1523/JNEUROSCI.1746-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haribabu B, Hook SS, Selbert MA, Goldstein EG, Tomhave ED, Edelman AM, et al. Human calcium-calmodulin dependent protein kinase I: cDNA cloning, domain structure and activation by phosphorylation at threonine-177 by calcium-calmodulin dependent protein kinase I kinase. EMBO J. 1995;14:3679–3686. doi: 10.1002/j.1460-2075.1995.tb00037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hart JR, Vogt PK. Phosphorylation of AKT: a mutational analysis. Oncotarget. 2011;2:467–476. doi: 10.18632/oncotarget.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang H, Bhat A, Woodnutt G, Lappe R. Targeting the ANGPT-TIE2 pathway in malignancy. Nat Rev Cancer. 2010;10:575–585. doi: 10.1038/nrc2894. [DOI] [PubMed] [Google Scholar]
- 54.Chang AC, Jellinek DA, Reddel RR. Mammalian stanniocalcins and cancer. Endocr Relat Cancer. 2003;10:359–373. doi: 10.1677/erc.0.0100359. [DOI] [PubMed] [Google Scholar]
- 55.Lowe KL, Navarro-Nunez L, Watson SP. Platelet CLEC-2 and podoplanin in cancer metastasis. Thromb Res. 2012;129(Suppl 1):S30–S37. doi: 10.1016/S0049-3848(12)70013-0. [DOI] [PubMed] [Google Scholar]
- 56.Li M, Mukasa A, Inda MM, Zhang J, Chin L, Cavenee W, et al. Guanylate binding protein 1 is a novel effector of EGFR-driven invasion in glioblastoma. J Exp Med. 2011;208:2657–2673. doi: 10.1084/jem.20111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Casazza A, Finisguerra V, Capparuccia L, Camperi A, Swiercz JM, Rizzolio S, et al. Sema3E-Plexin D1 signaling drives human cancer cell invasiveness and metastatic spreading in mice. J Clin Invest. 2010;120:2684–2698. doi: 10.1172/JCI42118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Gao J, Li L, Li Z, Du Y, Gong Y. The neuronal pentraxin II gene (NPTX2) inhibit proliferation and invasion of pancreatic cancer cells in vitro. Mol Biol Rep. 2011;38:4903–4911. doi: 10.1007/s11033-010-0632-y. [DOI] [PubMed] [Google Scholar]
- 59.Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song I, Huganir RL. Regulation of AMPA receptors during synaptic plasticity. Trends Neurosci. 2002;25:578–588. doi: 10.1016/s0166-2236(02)02270-1. [DOI] [PubMed] [Google Scholar]
- 61.Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 62.Manent JB, Jorquera I, Ben-Ari Y, Aniksztejn L, Represa A. Glutamate acting on AMPA but not NMDA receptors modulates the migration of hippocampal interneurons. J Neurosci. 2006;26:5901–5909. doi: 10.1523/JNEUROSCI.1033-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bodmer D, van den Hurk W, van Groningen JJ, Eleveld MJ, Martens GJ, Weterman MA, et al. Understanding familial and non-familial renal cell cancer. Hum Mol Genet. 2002;11:2489–2498. doi: 10.1093/hmg/11.20.2489. [DOI] [PubMed] [Google Scholar]
- 64.Ishiuchi S, Tsuzuki K, Yoshida Y, Yamada N, Hagimura N, Okado H, et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat Med. 2002;8:971–978. doi: 10.1038/nm746. [DOI] [PubMed] [Google Scholar]
- 65.Piao Y, Lu L, de Groot J. AMPA receptors promote perivascular glioma invasion via beta1 integrin-dependent adhesion to the extracellular matrix. Neuro Oncol. 2009;11:260–273. doi: 10.1215/15228517-2008-094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lyons SA, Chung WJ, Weaver AK, Ogunrinu T, Sontheimer H. Autocrine glutamate signaling promotes glioma cell invasion. Cancer Res. 2007;67:9463–9471. doi: 10.1158/0008-5472.CAN-07-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rzeski W, Ikonomidou C, Turski L. Glutamate antagonists limit tumor growth. Biochem Pharmacol. 2002;64:1195–1200. doi: 10.1016/s0006-2952(02)01218-2. [DOI] [PubMed] [Google Scholar]
- 68.Haas HS, Pfragner R, Tabrizi-Wizsy NG, Rohrer K, Lueftenegger I, Horwath C, et al. The influence of glutamate receptors on proliferation and metabolic cell activity of neuroendocrine tumors. Anticancer Res. 2013;33:1267–1272. [PubMed] [Google Scholar]
- 69.Prickett TD, Samuels Y. Molecular pathways: dysregulated glutamatergic signaling pathways in cancer. Clin Cancer Res. 2012;18:4240–4246. doi: 10.1158/1078-0432.CCR-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei C, Wang X, Chen M, Ouyang K, Song LS, Cheng H. Calcium flickers steer cell migration. Nature. 2009;457:901–905. doi: 10.1038/nature07577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei C, Wang X, Zheng M, Cheng H. Calcium gradients underlying cell migration. Curr Opin Cell Biol. 2012;24:254–261. doi: 10.1016/j.ceb.2011.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.