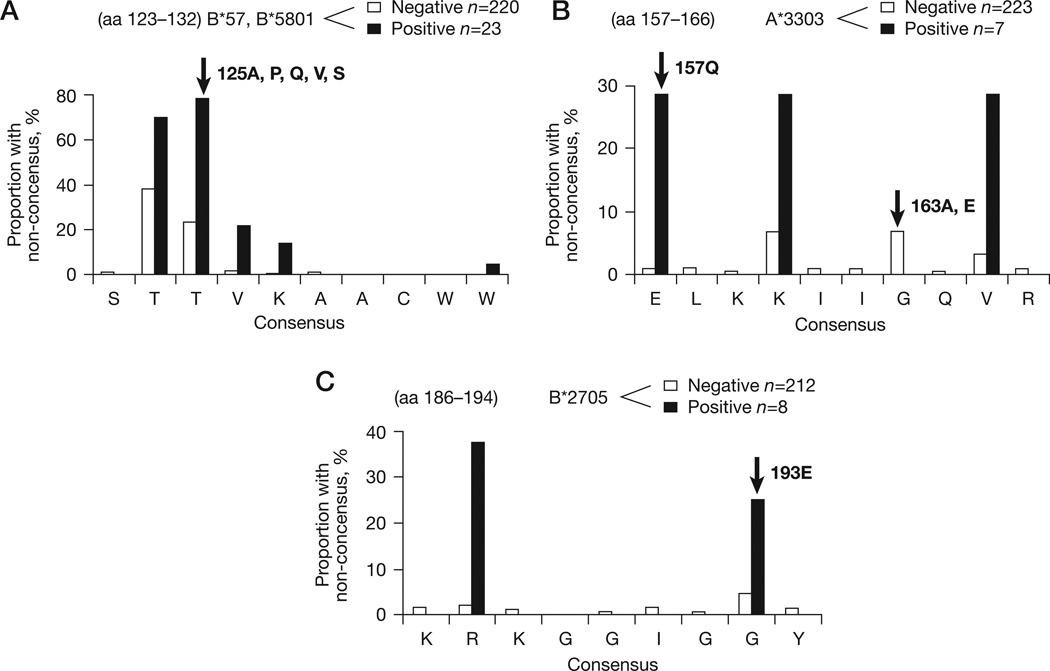
Figure 4. HLA allele-specific epitope variation at drug resistance sites.
(A) T125A is associated with HLA-B*57/*5801. (B) E157Q and G163A,E are associated with HLA-A*3303. (C) G193R is assoiciated with HLA-B*2705. Amino acid (aa) distribution at four codons (125, 157, 163 and 193, indicated with arrows) known to be integrase inhibitor resistance sites and associated with human leukocyte antigen (HLA) allele-specific polymorphism in reference data are shown. The polymorphism rate within and flanking the codons are shown in the presence (black bars) and absence (white bars) of the associated HLA allele. Polymorphism at 163 was associated with HLA-A*3303 in the reference associations dataset but not in the study population. Codon 72 was associated with HLA-A*0206 but is not included as only one individual carried this allele. HLA-B57/*5801-restricted SW10 and HLA-A*3303-restricted ER9 are published epitopes (Los Alamos National Laboratory), and KY9 is a predicted HLA-B*2705-restricted epitope based on the Epi-pred prediction programme [35]. Note the scale of the y-axis varies between panels.