Abstract
Summary: Aptamers are ‘synthetic antibodies’ that can bind to target molecules with high affinity and specificity. Aptamers are chemically synthesized and their discovery can be performed completely in vitro, rather than relying on in vivo biological processes, making them well-suited for high-throughput discovery. However, a large fraction of the most enriched aptamers in Systematic Evolution of Ligands by EXponential enrichment (SELEX) rounds display poor binding activity. Here, we present MPBind, a Meta-motif-based statistical framework and pipeline to Predict the Binding potential of SELEX-derived aptamers. Using human embryonic stem cell SELEX-Seq data, MPBind achieved high prediction accuracy for binding potential. Further analysis showed that MPBind is robust to both polymerase chain reaction amplification bias and incomplete sequencing of aptamer pools. These two biases usually confound aptamer analysis.
Availability and implementation: MPBind software and documents are available at http://www.morgridge.net/MPBind.html. The human embryonic stem cells whole-cell SELEX-Seq data are available at http://www.morgridge.net/Aptamer/.
Contact: RStewart@morgridge.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Aptamers are short, single-stranted DNA or RNA, which have the ability to specifically bind to a variety of targets including proteins (Ng et al., 2006) and the surface of cells (Daniels et al., 2003). Aptamers have gained significant interest as a promising alternative to antibodies because they are chemically synthesized, thermostable and can be readily produced using standard laboratory techniques. Importantly, aptamers can be distributed as sequence information rather than as a physical entity.
The generation of high-affinity aptamers typically starts with a random oligonucleotide pool. These oligonucleotides are then subjected to multiple rounds of in vitro target-based selection with polymerase chain reaction (PCR) amplification. This procedure is termed SELEX (Systematic Evolution of Ligands by EXponential enrichment). Several studies observed that the number of unique sequences decreases after target selection, suggesting that the decrease of sequence complexity is caused by the increase of sequence enrichment (Thiel et al., 2012). Also, ‘true-selected’ sequences were more likely to appear in multiple rounds if compared with non-selected sequences (Thiel et al., 2012). Thus, enrichment ratio-related methods are the most common approach to initially identify high-affinity aptamers. However, those methods suffer from a relatively high false-positive rate, where sequences are enriched but do not exhibit binding (Cho et al., 2010). This is likely because of PCR amplification bias.
To design aptamers that can bind to human embryonic stem cells (hESCs), we generated five rounds of hESC whole-cell SELEX sequencing data. We found that the validation rate is low (<50%) if we used the enrichment ratio-based method (data not shown). Prior work showed that PCR bias is a substantial confounding factor in aptamer analysis (Thiel et al., 2011). Therefore, there is a pressing need to have a computational approach that can accurately predict the binding potential of SELEX-derived aptamers without relying on aptamer read counts.
To this end, we developed MPBind, a novel statistical framework and pipeline to predict the binding potential of SELEX-derived aptamers. Our approach is based on the assumption that the binding potential of an aptamer can be broken down to the combination of binding of all n-mers (e.g. 6-mer) within a sequence. MPBind assesses all possible n-mers for metrics such as the relative frequency change and the relative abundance in the final round. Based on those observations, each n-mer is assigned a combined score. The binding potential of an aptamer is further inferred from the combination of all n-mers within the sequence. This approach integrates multiple moderately informative sources of data to generate high-confidence predictions, which is particularly important for Cell-SELEX where non-specific binding is prevalent.
2 METHOD
MPBind calculates four one-sided P-values for each motif, representing four different statistical tests for motif enrichment as described in Supplementary Methods. The P-values are then transformed to Z-scores (Z1, Z2, Z3 and Z4) via Z = Φ−1(1 − P), where Φ is the standard normal cumulative distribution function. For each motif, we used Stouffer’s method to combine the four Z-scores into one combined motif-level Z-score (Stouffer et al., 1949). We used an n-mer window to scan each aptamer for motifs to arrive at a Meta-Z-Score for the entire aptamer based on the aggregate Z-Score of all motifs within the aptamer (Supplementary Figure S1).
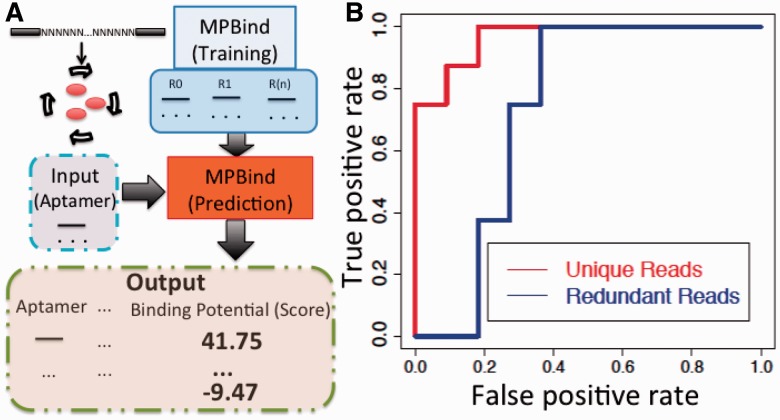
MPBind is implemented in Python/R and can be run in any Linux/Unix environment. As shown in Figure 1A, MPBind generates an MPBind training file from the high-throughput sequencing data from each round of SELEX. Then, statistical measures are calculated and MPBind computes the aptamer-level Meta-Z-Score, which is used to predict binding.
Fig. 1.
(A) Input, processing steps and output of MPBind. (B) ROC curves show MPBind trained on unique reads (AUC = 0.97) outperformed than that trained on redundant reads (AUC = 0.74). The motif length is set to 6 nt
3 RESULTS
We generated five rounds of whole-cell SELEX sequencing data using hESCs (Thomson et al., 1998). The initial library (R0) contains 21 M reads with ∼99% unique reads. PCR amplification by its nature generates redundant sequences. Therefore, if the sequencing depth is deep enough to cover all copies of reads in a pool, we should observe a drop in read complexity even without any selection or PCR bias. The read complexity in this scenario indicates the overall redundancy in a pool [Supplementary Figure S2 (A)]. However, if read depth is far less than all reads in a pool, as is typically the case for SELEX experiments, then the PCR cycles will not affect read complexity [Supplementary Figure S2 (B)]. This is because if there is no target selection and no PCR bias, PCR cycles will not change the relative abundance of each read species, and also because the sequencing depth is low compared with the number of available species; most species are sampled just once or not at all. Low read complexity then is likely indicative of some species being preferentially sampled. If a pool contains a fraction of favored sequences (by target selection or via PCR bias), with limited sampling depth, those favored sequences are repeatedly sampled and make the read complexity lower [Supplementary Figure S2 (C)]. To investigate the extent of incomplete sequencing in our pool, we compared the reads that overlap between sequencing runs in initial pool (R0) and Control-Seq (R1). The overlapping is minimal and only six reads are present in both sequencing runs [Supplementary Figure S2 (D)]. It indicates that our sequencing read depth is far from enough to cover all the reads in a pool. Incomplete sequencing of oligonucleotide pools is common in SELEX-Seq. This is because for a typical SELEX-Seq, the initial library pool requires at least ∼1010 to ∼1011 complexity to achieve the necessary diversity (Sassanfar and Szostak, 1993; Thiel et al., 2011). Our hESCs SELEX aptamers are 29 nt in length, and thus, the number of all possible sequences in the initial library pool is 429 (∼1017) in theory. Although the real number of sequences in the pool is likely to be much less than that, it is still far beyond the sequencing depth.
After one round of selection, the percentage of unique reads dropped to ∼51%. To investigate whether this drop in read complexity is a result of enrichment through true binding or PCR bias, we sequenced pools without target selection but with the same number of PCR cycles (Control-Seq). Our sequencing depth for our libraries was typically ∼107 sequences (Supplementary Table S1). As shown in Supplementary Table S1, the percentage of non-redundant reads in Control-Seq (R1) dropped to ∼47%. This drop in read complexity in the Control-Seq is likely due to PCR bias and likely indicates PCR bias in the SELEX-seq as well. For example, as shown in Supplementary Table S2, the top three enriched reads in the Control-Seq are ranked number 2–4 in terms of enrichment in the SELEX-Seq. It indicates that those enriched reads in SELEX-Seq are more likely due to being ‘selected by PCR’. PCR bias in SELEX-Seq has been previously observed (Thiel et al., 2011). To avoid PCR bias, we removed redundant reads from each round to train MPBind with parameter (n-mer = 6). The training set includes initial library (R0), SELEX-Seq (R1, R2, R3, R4 and R5) and Control-Seq (R5).
To evaluate the performance of MPBind, we selected 19 aptamers with a large dynamic range in their Meta-Z-Score (−45 to +47). We further defined binding as a notable shift of fluorescence intensity of cells bound by fluorescently labeled aptamer compared with controls (see Supplementary Figure S3 for details). The area under curve (AUC) of the receiver operating characteristic (ROC) curve of MPBind is 0.97. As expected, if we included redundant reads from each round to train MPBind, the AUC drops to 0.74, as shown in Figure 1B. The Spearman’s Rank correlation (Rho) between the predicted Meta-Z-Score (MPBind) and the binding potential (binding assay) is 0.79 [Supplementary Figure S3 (B)].
Aptamer 002 (Supplementary Table S3) is ranked as the top enriched aptamer in the final SELEX round (583 447 reads). The binding assay showed that this aptamer did not bind to hESCs. MPBind successfully predicts this to be a non-binding aptamer (Meta-Z-Score = −9.37). It is likely that the enrichment of Aptamer 002 is caused by PCR bias. This aptamer is also enriched in Control-Seq (R5) with 122 copies. However, the enrichment of this aptamer in Control-Seq is not as high as in SELEX-Seq, indicating that the extent of PCR bias can also be stochastic. This type of bias was also suggested by Dittmar et al. (2012), with the evidence that increasing copy number in SELEX-Seq did not necessarily display increasing binding affinity.
Aptamer 019 does not have sufficient read counts to support it as a high-affinity aptamer (no reads from R0 to R4 and only four reads in R5). However, MPBind predicted this to be a high-affinity aptamer (Meta-Z-Score = 9.01). The binding assay confirmed that this aptamer strongly binds to H1 cells [Supplementary Figure S3 (A) and Supplementary Table S3]. Interestingly, this indicates that MPBind can correctly predict aptamer affinity, even in the lack of read counts information (e.g. incomplete sequencing of oligonucleotide pools). In other words, MPbind is capable of achieving de novo predictions after sufficient training.
To examine how each individual statistic contributes to the prediction performance, we used Z1, Z2, Z3 and Z4 separately to predict aptamer binding potential. As shown in Supplementary Table S4, the AUCs of Z1, Z2, Z3 and Z4 ranged from 0.93 to 0.95, lower than when four Z scores are combined (AUC = 0.97). Thus, integrating multiple sources of information generates more confident predictions. To further investigate the impact of motif length on the prediction performance, we varied the motif length from 5 to 8 nt. As shown in Supplementary Figure S4, 6-mers achieved the best prediction performance.
To further evaluate the performance of MPBind, we tested it on another SELEX-Seq dataset (ESRP1) (Dittmar et al., 2012). Dittmar et al. generated five rounds of SELEX-Seq (ESRP1) data (R0, R2, R3, R6 and R7). For each round, we merged reads to unique reads (removed redundant reads) and trained MPBind with parameter n-mer = 6. Four aptamers (WT A, WT B, WT C and WT D) are selected by Dittmar et al. for binding validation using Electrophoretic Mobility Shift Assay (EMSA) analysis with increasing amounts of recombinant glutathione S-transferase (GST)-ESRP1 fusion protein (0–250 ng). Those four aptamers showed significant binding to ESRP1. As shown in Supplementary Table S5, our MPBind prediction showed that those four aptamers have Meta-Z-Scores: 32.8, 29.46, 37.3 and 35.59, respectively. To further confirm the binding, Dittmar et al., made 3–4 point mutations to each aptamer (as controls). The EMSA did not show significant binding for these mutant aptamers. The predicted Meta-Z-Scores for these mutant aptamers are −23.17, −11.54, −31.04 and 1.83, respectively (Supplementary Table S5). This indicates that MPBind can correctly predict aptamers that bind to ESRP1.
In summary, we show that MPBind is a useful tool for predicting binding aptamers from SELEX-Seq data and is robust to biases caused by PCR or by incomplete sequencing of aptamer pools. However, we should also note that the premise of MPBind is that the binding potential of an aptamer is dictated by the combination of n-mers. This assumption is valid for the two datasets we tested (hESCs whole-cell SELEX-Seq and ESRP1 protein SELEX-Seq). However, this might not be true for all SELEX-Seq data. In the future, we will further evaluate MPBind and continue to update it to be effective with a variety of SELEX-Seq datasets.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Krista Eastman for editorial assistance.
Funding: Funding for this research was provided by the Morgridge Institute for Research, the Institute for Collaborative Biotechnologies through the Army Research Office, the Garland Initiative at UCSB and NIH grant 1U54DK093467 (to H.T.S.).
Conflicts of interest: none declared.
REFERENCES
- Cho M, et al. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl Acad. Sci. USA. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DA, et al. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, et al. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell. Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EW, et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Stouffer SA, et al. 1949. The American Soldier: Adjustment During Army Life. (Studies in Social Psychology in World War II, Vol. 1.). Princeton University Press, Princeton, NJ. [Google Scholar]
- Thiel WH, et al. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PloS One. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel WH, et al. Nucleotide bias observed with a short SELEX RNA aptamer library. Nucleic Acid Ther. 2011;21:253–263. doi: 10.1089/nat.2011.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.