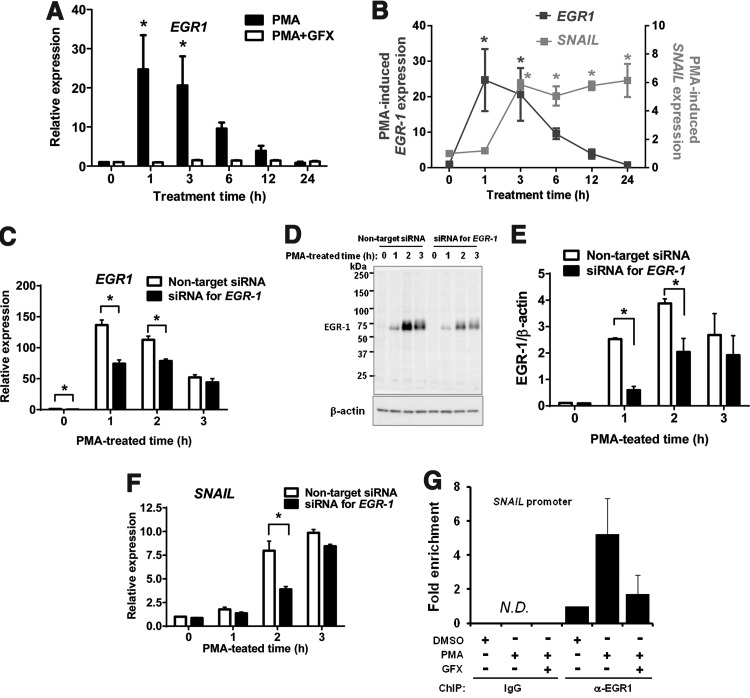
FIG. 3.
PMA-induced EGR-1 expression in hES cells. (A) qRT-PCR analysis of EGR-1 expression in the cells treated with PMA (black bars) or PMA together with GFX (white bars) for 24 h. (B) qRT-PCR analysis of EGR-1 (black square) or SNAIL (gray square) expression in the cells treated with PMA for 24 h. Expression levels were all normalized against GAPDH. The relative expression levels of each gene were calculated from the undifferentiated H9 hES cells untreated with PMA or PMA together with GFX. The data are represented as mean±SE (n=3). *P<0.05. (C) qRT-PCR analysis of PMA-induced EGR-1 expression in the cells transfected with siRNA targeting EGR-1 (black bars) or nontarget control (white bars). Cells were treated with PMA for 3 h. Expression levels were all normalized against GAPDH. The relative expression levels of EGR-1 were calculated from the nontarget siRNA-transfected H9 hES cells. The data are represented as mean±SE (n=3). *P<0.05. (D) Western blot analysis of PMA-induced EGR-1 expression in the cells transfected with siRNA targeting EGR-1 or nontarget control. Cells were treated with PMA for 3 h. (E) Quantification of the western blot result at (D). Cells were transfected with siRNA targeting EGR-1 (black bars) or nontarget control (white bars). The data are represented as mean±SE (n=3). *P<0.05. (F) qRT-PCR analysis of PMA-induced SNAIL expression in the cells transfected with siRNA targeting EGR-1 (black bars) or nontarget control (white bars). Cells were treated with PMA for 3 h. Expression levels were all normalized against GAPDH. The relative expression levels of SNAIL were calculated from the nontarget siRNA-transfected H9 hES cells. The data are represented as mean±SE (n=3). *P<0.05. (G) Chromatin immunoprecipitation (ChIP) assay. The cells were treated with PMA (10 nM) or PMA together with GFX (5 μM) for 1 h. N.D., not detected.