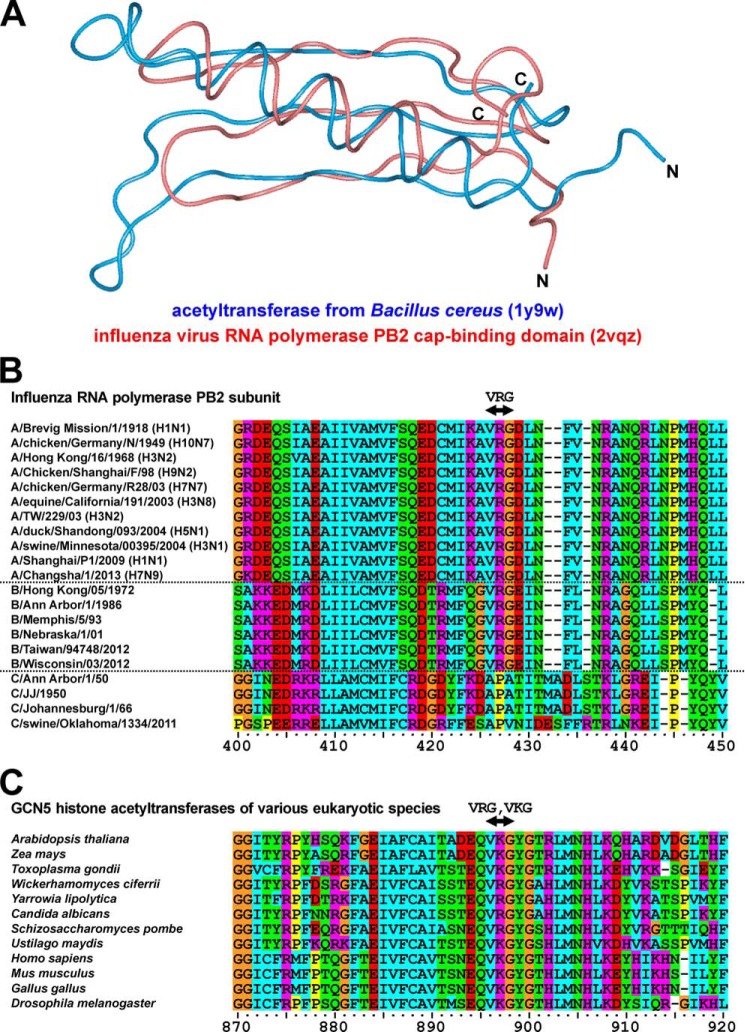
FIGURE 1.
Structural homology between PB2 and eukaryotic acetyltransferases. A, similarity of tertiary structures between the cap-binding domain of the PB2 subunit from the influenza A virus (the red ribbon; PDB code 2vqz) and that of the acetyltransferase from bacterium B. cereus (the blue ribbon; PDB code 1y9w). B, amino acid alignment of cap-binding domains of PB2 across various strains of influenza viruses. Sequences were placed in the order of years when each strain had emerged. Groups of influenza A, B, and C viruses were separated by dotted lines. The VRG site was highly conserved in strains of influenza A and B viruses. This site was not present in any strains of influenza C viruses. C, amino acid alignment of GCN5 acetyltransferases across various eukaryotic species. The VRG site and its corresponding sequence, the VKG site, are conserved in various GCN5 eukaryotic acetyltransferases. These amino acid sequences were aligned in a multiple-sequence alignment program, ClustalX. Orange, glycine; yellow, proline; cyan, hydrophobic neutral amino acids; green, hydrophilic neutral amino acids; red, acidic amino acids; magenta, basic amino acids.