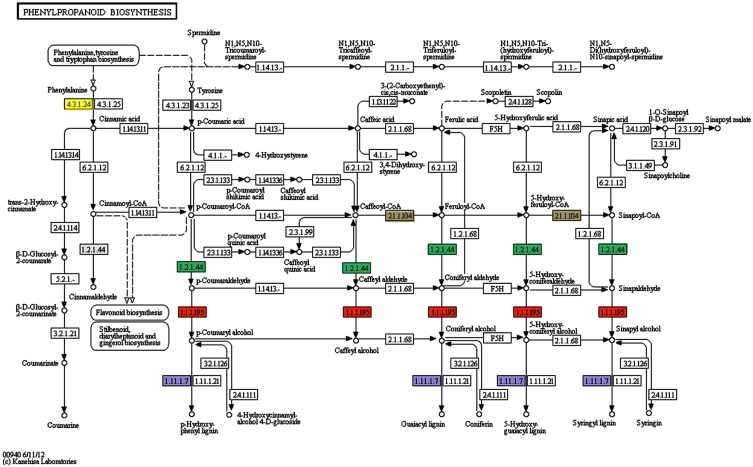
Figure 3.
Patway map from KEGG for the phenylpropanoid biosynthesis pathway. Several genes involved in this pathway have been found to be induced in olive roots (Schilirò et al., 2012) and/or aerial tissues (this study) upon Pseudomonas fluorescens PICF7 root inoculation. The enzymes reductase (1.2.1.44) (green rectangles) induced in aerial tissues, the cinnamyl-alcohol deydrogenase (1.1.1.95) (orange rectangles), the lactoperoxidase (1.11.1.7) (violet rectangles), the phenylalanine ammonia-lyase (4.3.1.5)* induced in roots and aerial tissues, and the caffeoyl-CoA Omethyltransferase (2.1.1.104) (gray rectangles) induced in roots, are mapped. *Phenylalanine ammonia-lyase (EC4.3.1.5) is now divided into three different enzymes: EC4.3.1.23 (tyrosine ammonia-lyase), EC4.3.1.24 (phenylalanine ammonia-lyase), and EC4.3.1.25 (phenylalanine/tyrosine ammonia-lyase) according to IUBMB Enzyme Nomenclature (http://www.chem.qmul.ac.uk/iubmb/enzyme/EC4/3/1/5.html). Yellow rectangle is phenylalanine ammonia-lyase.