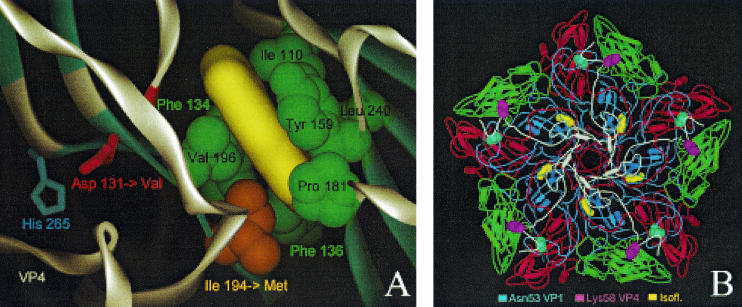
FIG. 1.
Localization of drug-resistant and -dependent mutations and modeling of drug binding. (A) Close-up of the 3(2H)-isoflavene binding site near VP1 Ile194. The 3(2H)-isoflavene ligand (yellow) is modeled into the bottom of the hydrophobic pocket formed by VP1 Ile110, Phe134, Phe136, Tyr159, Pro181, Ile194, Val196, and Leu240. Mutation of Ile194 into a longer Met side chain (indicated by an orange arrow) partially fills the bottom of the pocket, abolishing efficient binding of the isoflavene. Also shown is VP1 Asp131 (red), a second mutation (Asp→Val) that confers drug resistance. Asp131 forms contacts with VP4 (through His265) and the backbone (NH of Tyr198) of one of the beta strands that is part of the pocket wall. In particular, loss of the latter contact could induce a local conformational change of the Val196-Pro197-Tyr198 backbone segment (highlighted in red), leading to a change in shape and, possibly, also leading to a change in the accessibility of the pocket. (B) The locations of the two mutations resulting in a drug-dependent phenotype, VP1 Asn53 to Ser and VP4 Lys58 to Glu, are shown in the context of a VP1 to VP4 pentamer seen from inside the capsid (VP1, blue; VP2, green; VP3, red; VP4, white). Both residues lie on the inner surface of the capsid and are far from the isoflavene-binding pocket (yellow).