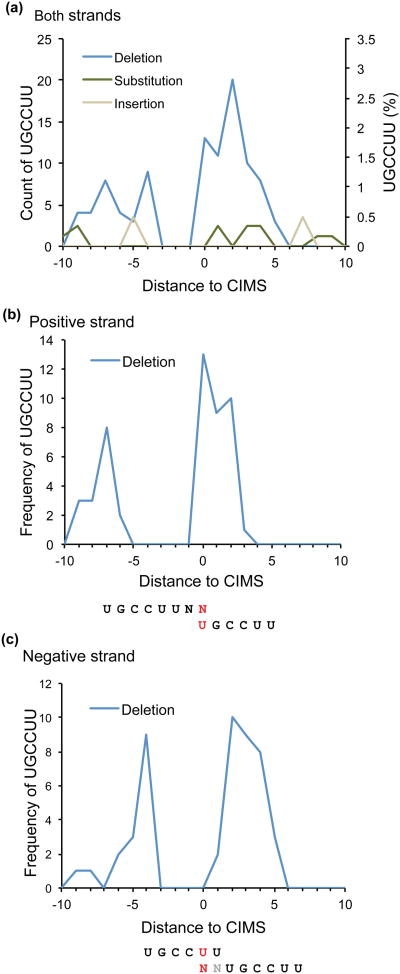
Fig. 6. Enrichment of miR-124 seed site matches in sequences [-10 nt, +10 nt] around robust CIMS (FDR≤0.001) (steps 110-11).
(a) The frequency of miR-124 seed matches (UGCCUU) relative to the position of reproducible deletion sites is plotted. The percentage of sites with miR-124 seed matches is shown on the vertical axis. Crosslinking predominantly occurs in positions immediately flanking the miR-124 seed match sequences. (b,c) Frequency of miR-124 seed matches relative to the position of reproducible deletion sites is plotted on positive (a) and negative (b) strands. Ambiguity in assigning the position of nucleotide deletions arises when crosslinking occurs in a stretch of the same nucleotide; novoalign assigns the deletion to the last position in the stretch relative to the positive strand. Therefore, CIMS on both strands are examined separately. The bimodal distribution of deletions is more evident when transcripts on both strands are separated. Below the graphs, the most frequent positions of miR-124 seed match sequences (UGCCUU, black) and the most frequently deleted nucleotides (red) are highlighted in panels (b) and (c).