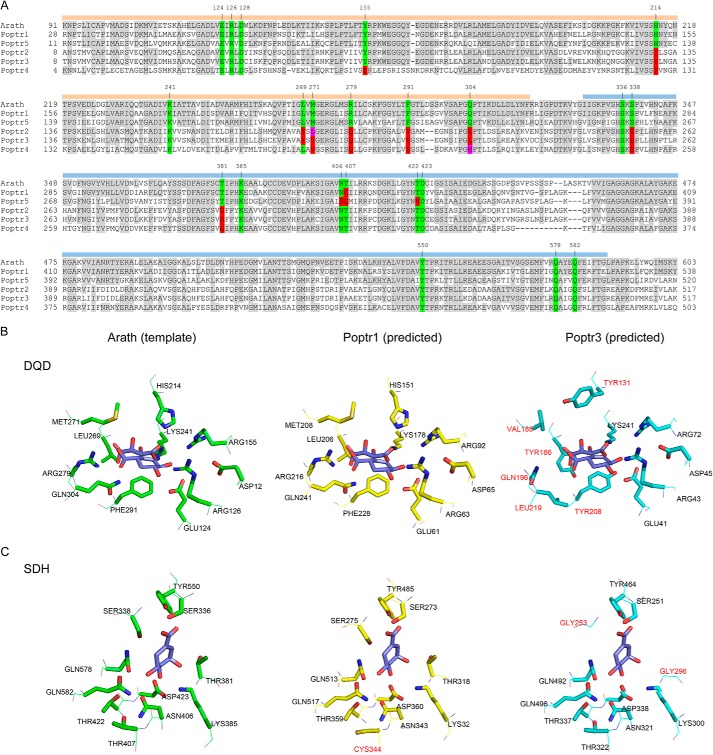
FIGURE 2.
Alignment of DQD/SDHs from P. trichocarpa (Poptr1 to Poptr5) and A. thaliana (A) and active site structure predictions of Poptr1 and Poptr3 in comparison with the known DQD/SDH structure from Arabidopsis (Protein Data Bank code 2O7Q) (B and C). A, shown is the alignment as used by Phyre for structure prediction. Note that the N-terminal signal peptide was not included in the Arabidopsis structure determination by Singh and Christendat (53) and was thus omitted from the alignment. Amino acids identical to the Arabidopsis template are shaded in gray. Active site residues of the Arabidopsis protein are highlighted in green. Identical amino acids at the respective position in the poplar sequences are also highlighted in green, and different amino acids are contrasted in red or magenta. Numbers above the sequence indicate the Arabidopsis amino acid position. The two functional domains (DQD and SDH) are highlighted by orange and blue bars, respectively. B, structure models were predicted with Phyre; DQD active sites (B) and SDH active sites (C) were visualized as stick models with PyMOL. Amino acids in the Poptr1 or Poptr3 predictions differing from the Arabidopsis template structure are highlighted in red.