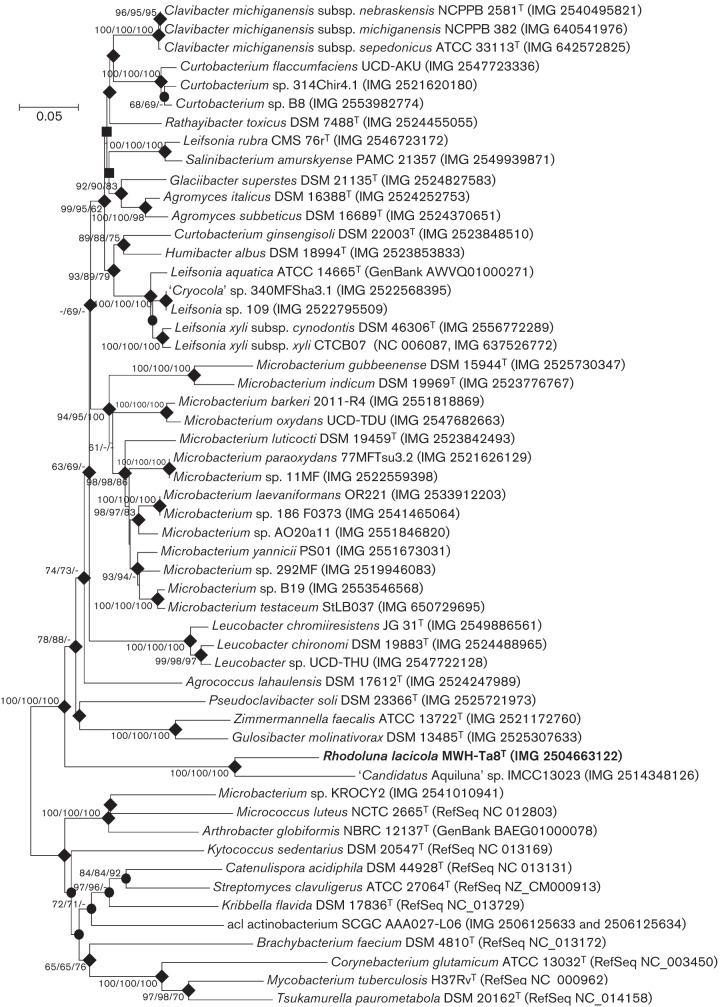
Fig. 2.
NJ tree based on partial RpoB (DNA-directed RNA polymerase subunit beta) amino acid sequences reconstructing the phylogenetic position of strain MWH-Ta8T. Amino acid sequences were used for phylogenetic analysis in order to omit phylogenetic noise potentially cause by the pronounced differences in DNA G+C content of rpoB genes (range 51 to 68 mol%). Results from analyses by the ML and MP methods are also indicated. Bootstrap values (percentage of replicates) above the threshold of ≥60 % are shown for those nodes supported at least in inference by one of the three methods; values are presented in the order NJ/ML/MP. Filled diamonds indicate nodes reconstructed by all three methods independent of their respective bootstrap values; filled circles indicate those nodes only present in NJ and ML trees; filled squares indicate nodes only reconstructed by the NJ and MP methods. If available, IMG Gene ID numbers of genes encoding the RpoB proteins are given in parentheses; for other proteins, the RefSeq or GenBank accession numbers are provided. Microbacterium sp. KROCY2 represents a strain classified by the IMG database as a member of the family Microbacteriaceae, and acI actinobacteria SCGC AAA027-L06 represents a freshwater actinobacterium affiliated with the acI lineage (Garcia et al., 2013). Ten type strains of species not affiliated with the family Microbacteriaceae were used as an out-group. Bar, 0.05 substitutions per amino acid position.