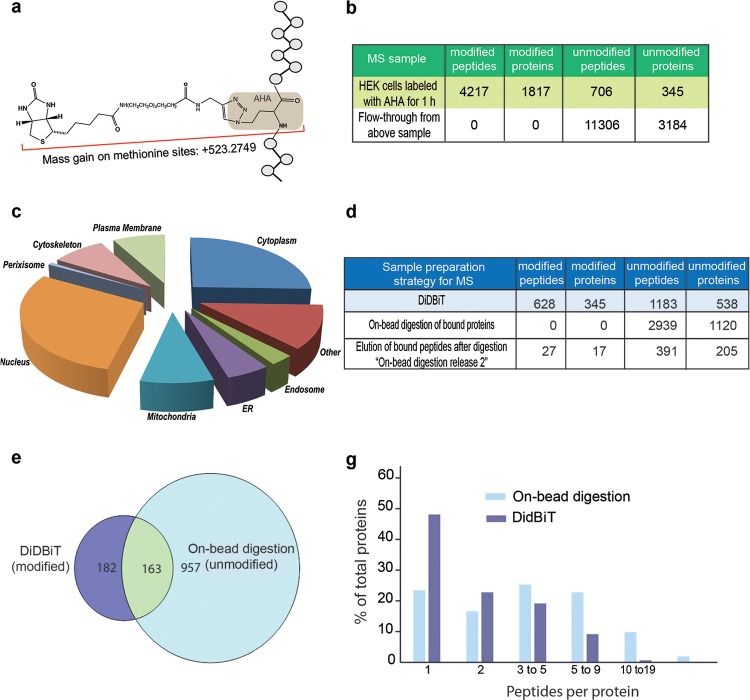
Figure 4.
Application of DiDBiT to identify newly synthesized proteins in HEK cells. (a) Schematic of the biotin-AHA modification with mass gain of 523.2749 on methionine sites in peptides. (b) HEK cells were exposed to AHA for 1 h. Starting from 10 mg of protein lysate and using MudPIT analysis, we identified 4210 modified peptides corresponding to 1817 newly synthesized proteins by a mass gain of 523.2749. We identified and filtered out 711 unmodified peptides corresponding to 345 proteins. As expected, only unmodified peptides were detected in the analyses from NeutrAvidin beads flow-through after peptide enrichment, no modified peptides were detected. (c) Analysis of the cellular compartments from which newly synthesized proteins were identified (d) Comparison of DiDBiT and on-bead digestion to detect AHA-biotin labeled newly synthesized proteins from HEK cells. Starting from 6 mg of protein lysate and using reverse-phase separation coupled to MS analysis, DiDBiT increased detection of modified proteins 23-fold. (e) Venn diagram showing the selective identification of biotinylated proteins using DiDBiT compared to the detection of unmodified peptides from on-bead digestion. More than half of the biotinylated proteins identified by DiDBiT were not detected in samples from on-bead digestion. The majority of unmodified proteins (86%) detected by on-bead digestion were not detected by with DiDBiT, and are likely contaminants from incomplete purification of biotinylated proteins. (g) Plots of the number of peptides per protein for DiDBiT and on-bead digestion. AHA-labeling results in relatively sparse coverage.