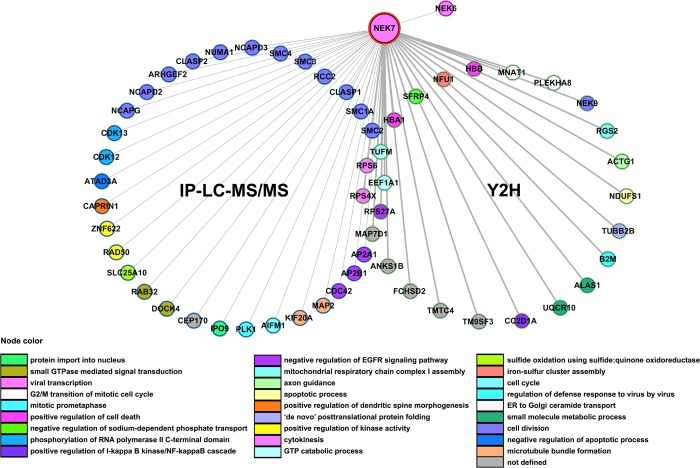
Figure 1.
Interaction network of human NEK7 protein partners identified by IP-LC–MS/MS and Y2H system screens. Two circles represent the interactions retrieved by both methods. Y2H interactors also retrieved by IP-MS/MS are depicted in the intersection between the two circles. Thicker edges correspond to direct interactions retrieved by the Y2H screens. The proteins’ color code refers to their biological function given by the top enriched Gene Ontology biological processes (p ≤ 0.05). The proteins included in these interaction analyses were selected from the IP-LC–MS/MS by the following criteria: exclusive peptides (no peptides in replicated FLAG-control), overlapping proteins with Y2H and proteins not deposited in the Contaminant Repository for Affinity Purification (CRAPome). NEK9 (also identified by the Y2H screening) and NEK6 were already described to interact with NEK7.7,39 The protein–protein interaction network was built using the Integrated Interactome System (IIS) platform26 and visualized using the Cytoscape software.27