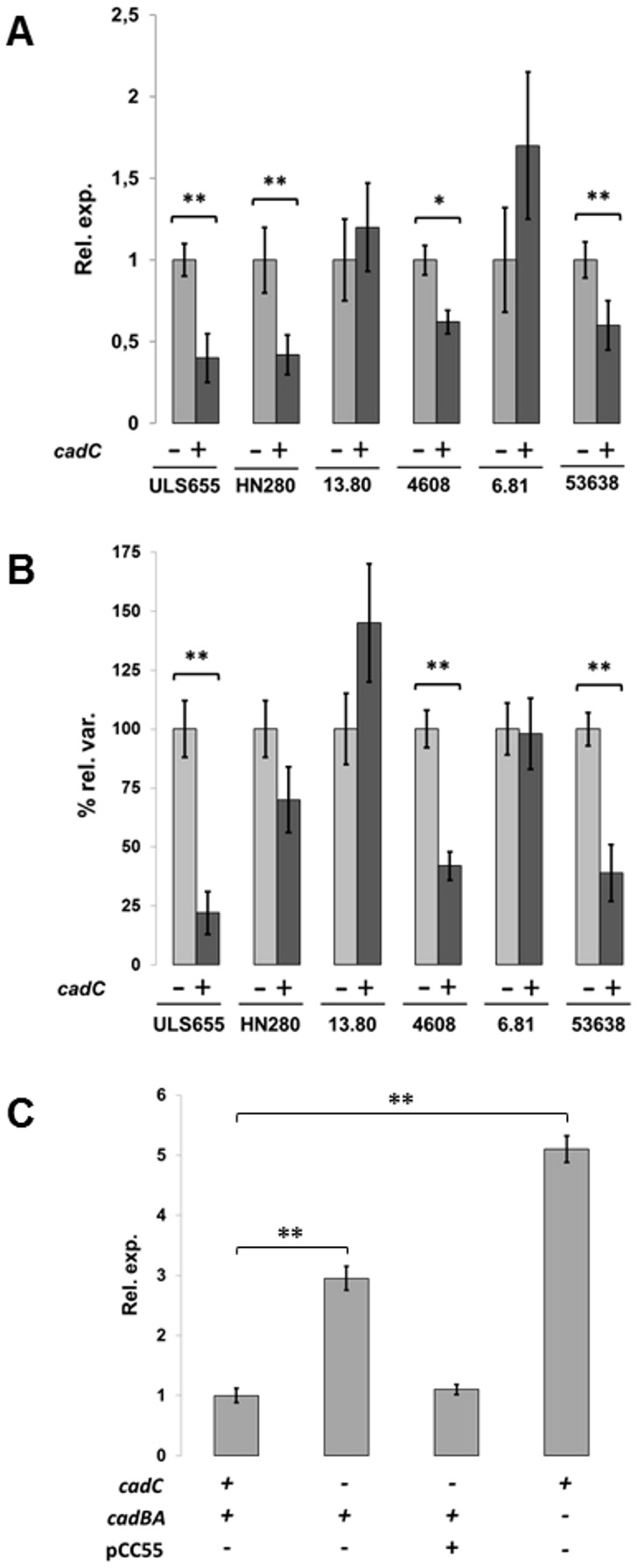
Figure 5. Cadaverine interferes with speC expression.
Experiments were performed using strains lacking only the cadC gene (EIEC HN280, 4608 and 53638 and E. coli K-12 ULS655) or the entire cad operon (EIEC 13.80 and 6.81). Strains were transformed with pCC55 (a pACYC184 derivative containing a functional cadC gene) or pACYC184. (A) Transcription of speC in the presence or absence of cadaverine as monitored by Real-Time PCR. At least three wells were run for each sample and the error bars display the calculated maximum (RQMax) and minimum (RQMin) levels that represent standard error of the mean expression level (RQ value). * denotes 0.05>p≥0.01; ** denotes p<0.01 (B) Ornithine decarboxylase (ODC) activity in the presence or absence of cadaverine. ODC activity was measured in total cell extracts by assaying putrescine production. The synthesized putrescine was made fluorescent through chemical modification and separated by TLC. The fluorimetric data were normalized against the total protein content of each sample. Results are shown as variation (percentage) in strains carrying pCC55 (cadC) vs the corresponding wt strain carrying the backbone construct. * denotes 0.05>p≥0.01; ** denotes p<0.01. (C) Transcription of speC in E. coli K-12 (MG1655); ** denotes p<0.01. Transcription in strains producing or lacking cadaverine was monitored by Real-Time PCR as in panel A.