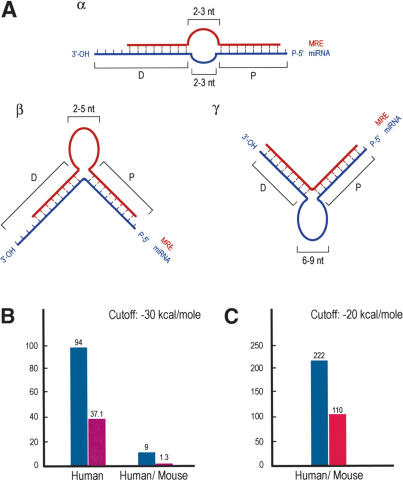
Figure 5.
MicroRNA Binding Rules and Statistics. (A) Schematic representation of miRNA:MRE (target mRNA) bindings (miRNA binding rules). (Blue) miRNAs; (red) MRE; (P) proximal (relative to 5′-end of miRNA) region of miRNA:MRE binding; (D) distal region of binding. (Panel α) Loop, length (on each sequence) = 2–3 nt. (Panel β) Single MRE central bulge, length = 2–5 nt. (Panel γ) Single miRNA central bulge, length = 6–9 nt. Proximal binding characteristics are ≥7-nt base pairing between miRNA and MRE; the 5′-most nucleotide of the miRNA may or may not base pair with MRE; one symmetric single nucleotide bulge allowed (i.e., the single nucleotide bulge is surrounded by an equal number of base-paired nucleotides). Distal binding characteristics are ≥5-nt base pairing between miRNA and MRE; nucleotide bulges allowed. The last (toward the 3′ end) nucleotides of the miRNA may or may not base pair with the MRE. (B) Hits between 10 human miRNAs (blue bar) or shuffled RNAs (purple bar) and the 3′-UTR database of annotated human mRNAs or the conserved human/mouse 3′-UTR database (initial analysis). (C) Hits between 94 human miRNAs (blue bar) or shuffled controls (with the same compositional properties as the authentic miRNAs; red bar) and the conserved human/mouse 3′-UTR database extracted using EnsMart.