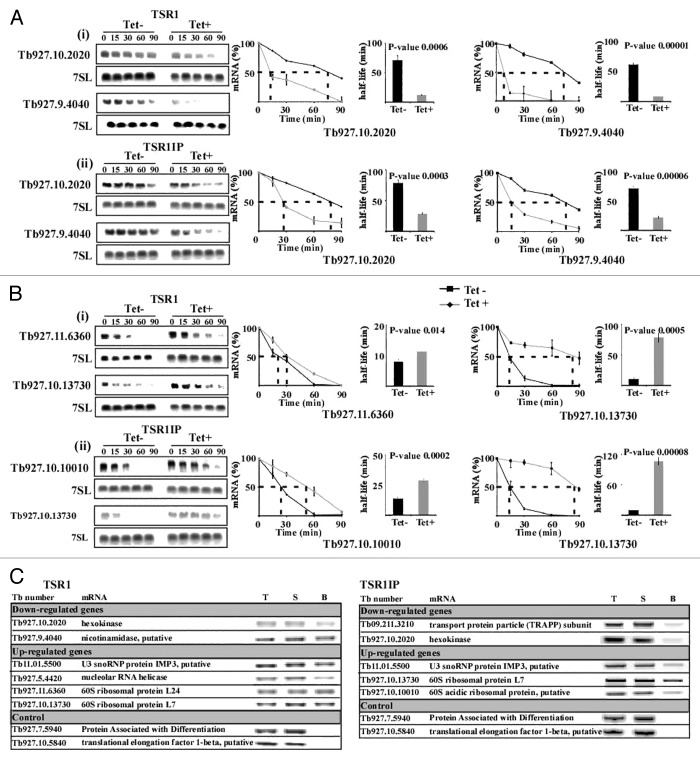
Figure 3. Changes in stability of mRNAs upon silencing of TSR1 and TSR1IP. Uninduced and TSR1 or TSR1IP silenced cells (3 d after induction) were treated with sinefungin (2 µg/ml) and, after 10 min, with Actinomycin D (30 µg/ml). RNA was prepared at the time points indicated above the lanes, separated on a 1.2% agarose-formaldehyde gel, and subjected to northern analysis with the indicated gene-specific probes. 7SL RNA was used to control for equal loading. (A) The half-life of downregulated transcripts. (i), TSR1; (ii), TSR1IP. (B) Half-life of upregulated genes. (i) TSR1; (ii) TSR1IP. The hybridization signals were measured by densitometry. The decay curves are shown with the blots, and the half-life is illustrated by the broken lines. The decay in the absence of induction (-Tet) is indicated by a black line, and following induction (+Tet) by a gray dashed line. The experiments were repeated three times; each data point corresponds to the average, with the standard deviation indicated. The half-life was then calculated by fitting the normalized RNA levels to an exponential decay. The half-lives (averaged over the three experiments) are shown as bars with standard deviations, along with P values (t test) for the difference between the half-lives in uninduced compared with silenced cells. (C) Affinity selection of TSR1 and TSR1IP substrates using tagged proteins. Whole cell extracts were prepared from 5 × 109 cells with PTP tagged TSR1 and TSR1IP after 5 min of UV cross-linking, and the extract was subjected to affinity purification on IgG beads, as described previously.27 RNA was eluted from the beads, and cDNA was prepared from the bead eluate, 5% of the total RNA, and 5% of the RNA from the supernatant. cDNA was subjected to PCR with SL forward primer and reverse primer from the ORF of each gene (specified in Supplemental S-1). As a control, two transcripts whose levels were unchanged in the silenced cells were used. T, total RNA; S, supernatant; B, beads.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.