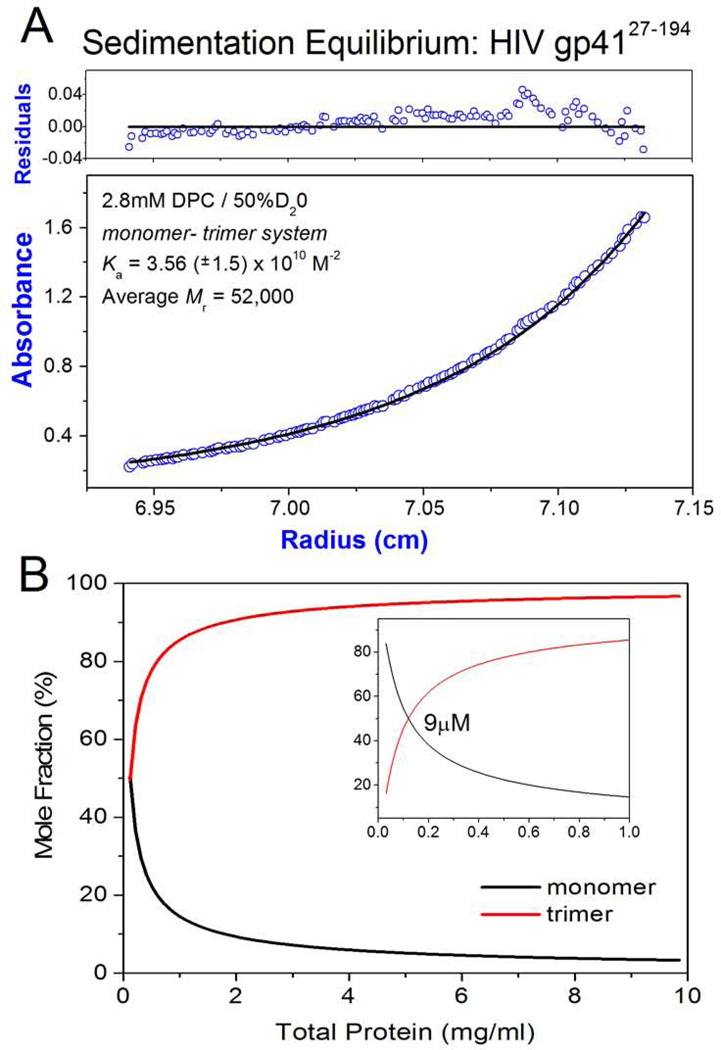
Figure 4. Analytical Ultracentrifugation.
a) Sedimentation equilibrium ultracentrifugation of HIV-1 gp4127–194: Panels are absorbance (bottom panel) and residuals (upper panel). Opened circles show 280nm absorbance gradients in the centrifuge cell. The solid line indicates the calculated fit for the monomer-trimer association. Residuals show the difference in the fitted and experimental values as a function of radial position. The data shown refers to a 15N13C2H labelled protein with a calculated monomeric mass of 22,000 (sample used directly for NMR analyses). The concentration range of the gradient (open circles) at equilibrium is ~ 1.0 µM – 20 µM. b) The monomer – trimer potential of gp4127–194 is indicated as mole fraction of monomer and trimer plotted as function of total protein concentration. Profiles were constructed using the Ka value indicated in (A). The insert is at a tenfold lower protein concentration and the indicated protein molarity corresponds to ~ 50% monomer and 50% trimer composition. The molecular weight of the non-labelled gp4127–194 construct is 19.65 kDa (1mg/ml = 50.9µM) and the NMR measurements described in the study were performed at ~ 20mg/ml (1mM).