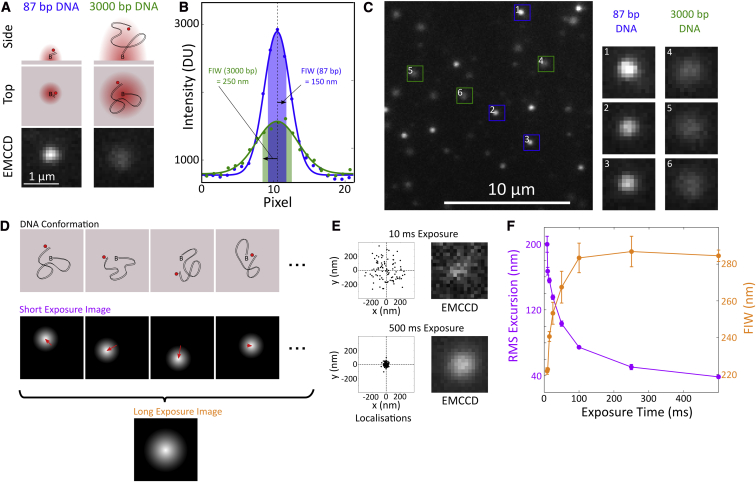
Figure 1.
Principle of TFM. (A) Cartoon and images of single emitters. A Cy5 fluorophore, shown as a red circle was attached to DNA and tethered to a cover slip using a biotin-neutravidin interaction. An 87 bp DNA results in a diffraction limited image (red in the cartoons); but a 3000 bp DNA allows the fluorophore to diffuse about the tether point during a frame, causing the image to appear broader. The bottom panels show EMCCD data. (B) Gaussian fits to the pixel intensity values, in digital camera units (DU), from cross-sections through the EMCCD data shown in (A) demonstrate the difference in FIW (highlighted region and arrow). (C) Example regions of interest corresponding to 87 and 3000 bp DNA are shown in the right panels. A section of our field of view: 87 bp DNA appear as bright, narrow spots; and 3000 bp DNA appear as dimmer, broader spots. (D) Cartoon of two different imaging schemes. Using short exposures, we see a diffraction-limited FIW and the DNA length is apparent as a mean excursion (red arrows). Using long exposures, the averaging is done during the camera acquisition and we see an increased FIW, fixed in the image plane. (E) Single-molecule localizations from sequential frames of a 100-frame movie of 3000 bp DNA, taken with 10 or 500 ms exposure times. Localizations (i.e., the position of the peak of the fitted Gaussian) are shown in black, and the mean of all localizations is indicated by the dashed lines. Representative images at the two exposure times are shown. The brightness of the images has been adjusted because the 500 ms exposure gives a much brighter image. (F) FIW and root mean square (RMS) excursion for 3000 bp DNAs at frame times from 8.5 to 500 ms. There is a decrease in FIW and increase in mean excursion as the frame time becomes comparable with the DNA relaxation time. Error bars are the standard error in mean from five molecules at each exposure time. Localization error is less than 10 nm in all cases.