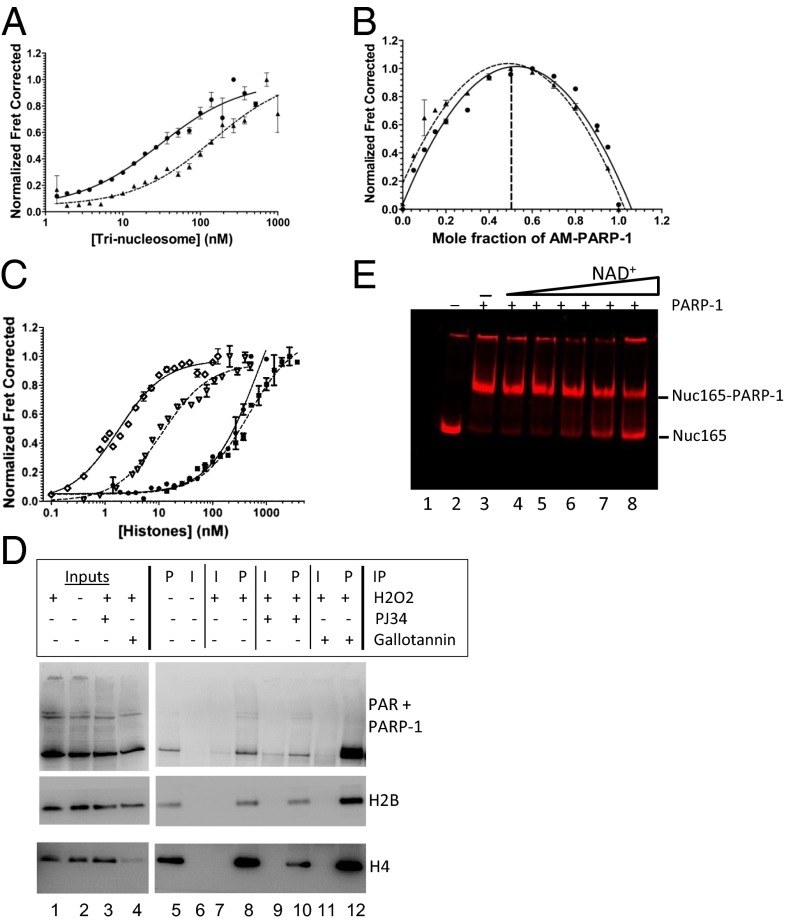
Fig. 2.
PARylation converts PARP-1 from a chromatin architectural protein to a histone chaperone. (A and B) Representative binding curves and stoichiometry measurements of AM–PARP-1 to LE-Tri (●, solid lines) and NLE-Tri (▲, dashed lines). (C) Binding curves for PARP-1 and AM–PARP-1 to histones. H2A-H2B–PARP-1 (●, solid line), H3-H4–PARP-1 (■, dashed line), H2A-H2B–AM–PARP-1 (♢, solid line), and H3-H4–AM–PARP-1 (▿, dashed line) are shown. (D) Endogenous PARylated PARP-1 coimmunoprecipitates soluble histones. U2OS cells were treated with H2O2 in combination with PJ34 or gallotannin as indicated. PAR antibodies (P) or control IgG (I) were used in immunoprecipitation (IP) assays from soluble lysates. Bound proteins were detected by immunoblotting with a mixture of PARP-1 and PAR antibodies (Top) and H2B and H4 antibodies (Middle and Bottom). (E) AM–PARP-1 does not disassemble nucleosomes. Labeled Nuc165 with PARP-1 and 30Nick DNA was incubated with increasing amounts of NAD+. Samples were analyzed by native PAGE and visualized by gel FRET between H2B (cyanine 5 633/670) and H4 (Alexa488 488/520). Lane 2 contains nucleosomes alone; lane 3 contains nucleosomes with PARP-1 and 30Nick DNA; and lanes 4–8 contain nucleosomes with PARP-1, 30Nick DNA, and 0.1, 1, 10, 20, and 40 μM NAD+, respectively. FRET between histones H2B and H4 is observed in all lanes, indicating that nucleosomes remain intact throughout.