Fig. 3.
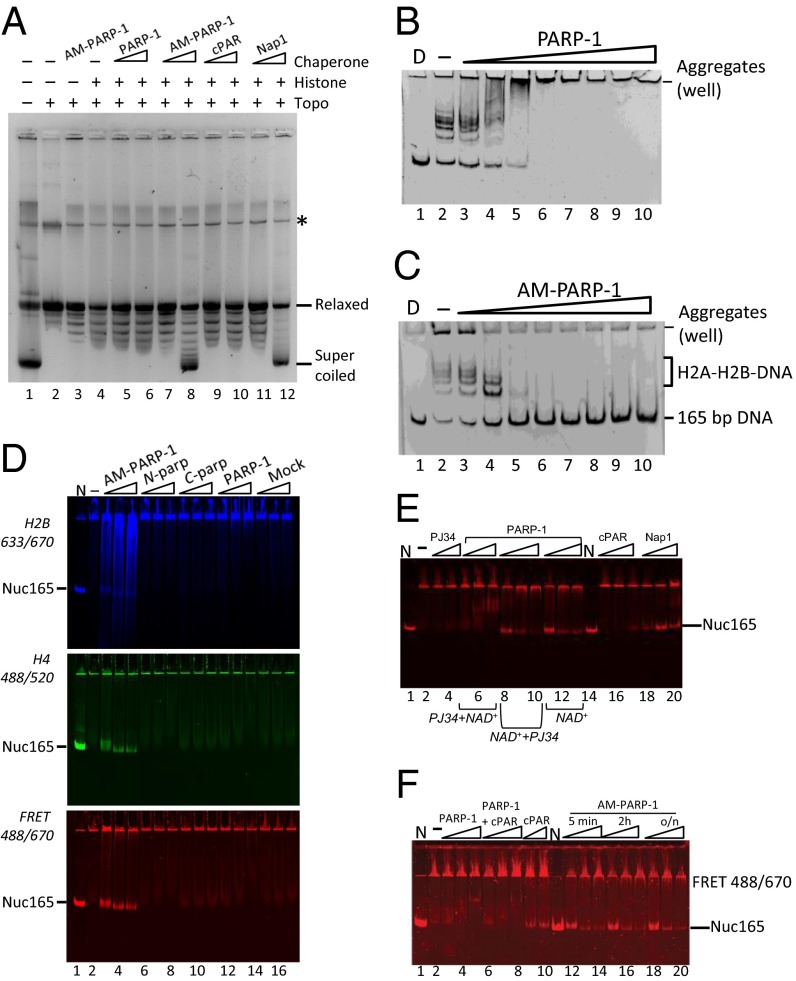
AM–PARP-1 is a nucleosome assembly factor. (A) AM–PARP-1 mediates plasmid supercoiling. Lane 1 contains pGEM-3Z plasmid before relaxation. Lane 2 contains relaxed plasmid. Lane 3 contains 200 nM AM–PARP-1 and no histone octamer. Lanes 4–12 contain 40 nM histone octamer. Lanes 5 and 6 contain 40 and 200 nM unmodified PARP-1. Lanes 7 and 8 contain 40 and 200 nM AM–PARP-1. Lanes 9 and 10 contain 40 and 200 nM cPAR. Lanes 11 and 12 contain 40 and 400 nM Nap1. The asterisk (*) indicates a contaminant present in the DNA. Topo, topoisomerase. (B and C) AM–PARP-1, but not unmodified PARP-1, removes H2A-H2B from DNA. Lane 1 contains D only, lane 2 contains 165-bp DNA with a 7 M excess of H2A-H2B. Lanes 3–10 contain 10, 50, 120, 240, 320, 500, 750, and 1,000 nM unmodified PARP-1 or AM–PARP-1. (D) RAC assay, in which 10 nM 165-bp DNA was incubated with 10 nM (H3–H4)2 tetramer (donor) and 120 nM H2A–H2B dimer (acceptor). An increasing amount of PARP-1 was added, and samples were analyzed by 5% native PAGE, followed by visualization of H2B fluorescence (Top), H4 fluorescence (Middle), and FRET between H2B and H4 (Bottom). Lane 1 contains fluorescently labeled nucleosome (N). Lane 2 contains no PARP-1. Lanes 3–5 contain 10, 50, and 120 nM AM–PARP-1 (0.083, 0.42, and 1 M ratio of AM–PARP-1 to H2A–H2B dimer). The same amounts of N-parp, C-parp, unmodified PARP-1, and a mock reaction without PARP-1 were added as indicated (lanes 15–17). (E) Nucleosome assembly activity of AM–PARP-1 cannot be recapitulated by PAR chains and does not require AM–PARP-1 to be enzymatically active. The assay was done as in D; only the FRET scan is shown here. Lanes 1 and 14 contain N. Lane 2 contains DNA, (H3–H4)2 tetramer, and excess H2A–H2B (as in A, lane 2). Lanes 3 and 4 contain 50 and 120 μM PJ34 inhibitor. Lanes 5–7 contain 10, 50, and 120 nM PARP-1 preincubated with a final concentration of 1 mM PJ34 before addition of NAD+ (PJ34 + NAD+). Lanes 8–10 contain 10, 50, and 120 nM PARP-1 preincubated with NAD+ before addition of PJ34 (NAD+ + PJ34). Lanes 11–13 contain 0, 50, and 120 nM PARP-1 together with NAD+ (no inhibitor). Lanes 15–17 contain 10, 50, and 120 nM cPAR. Lanes 18–20 contain 10, 50, and 120 nM Nap1. (F) Length of automodification reaction does not affect assembly activity. The assay was done as in D; only the FRET channel is shown. Lanes 1 and 11 contain N. Lane 2 is DNA, (H3–H4)2 tetramer, and excess H2A–H2B as in D (lane 2). To this sample, 10, 50, and 120 nM PARP-1 (lanes 3–5); PARP-1 together with PAR (lanes 6–8); PAR alone (lanes 9–10); and AM–PARP-1 modified for 5 min (lanes 12–14), 2 h (lanes 15–17), or overnight (lanes 18–20) were added (automodification is shown in Fig. S3A, lanes 15–18).