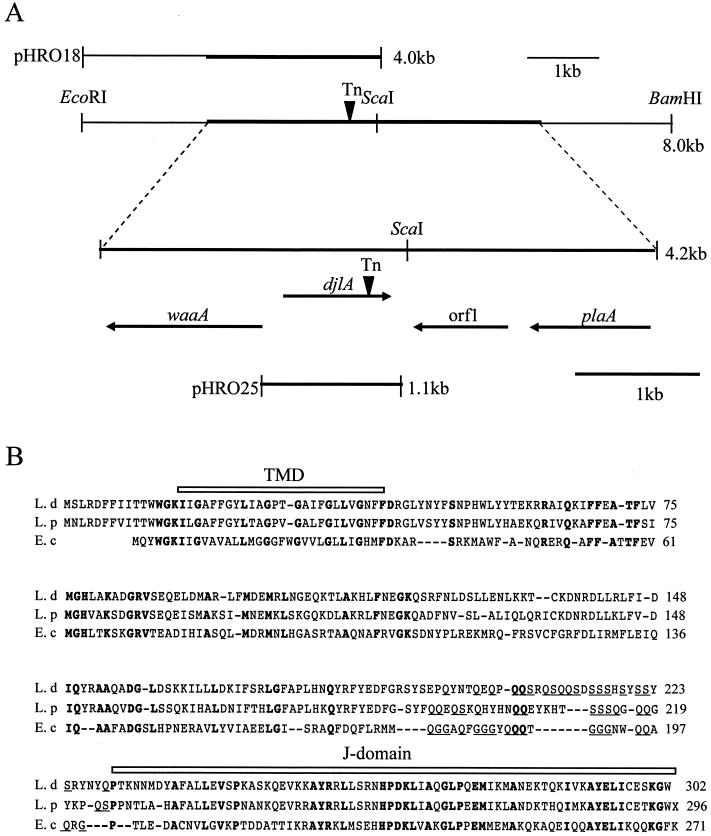
FIG. 3.
Chromosomal arrangement of the region surrounding the djlA gene and sequence alignment of DjlA proteins. (A) At the top is a plasmid used for complementation studies (pHRO18) and an 8-kb region of the L. dumoffii cosmid clone including the djlA gene, along with the location of relevant restriction enzyme sites. The thick line represents the DNA region that we sequenced. Below these diagrams, the distance between the djlA gene and neighboring genes and the orientation and size of the transcribed genes are delineated by the arrows below the 4.2-kb sequenced region. Another plasmid used for complementation studies (pHRO25) is also shown. The site of the Tn903dIIlacZ insertion (Tn) is indicated by the inverted arrowhead. The full names of the gene mapped are as follows: waaA, Kdo transferase gene; djlA, dnaJ-like A gene; plaA, lysophospholipase A gene. Orf1 is a putative open reading frame which showed no homology to known genes. (B) Sequence similarity of the predicted Dj1A protein of L. dumoffii (L.d, top line), L. pneumophila (L.p, middle line) and E. coli (E.c, bottom line). Amino acid residues conserved in the three sequences, appear in bold type. Gaps marked by dashes are introduced to reveal the maximal similarity among the sequences. The C-terminal J-domain and the N-terminal TMD are shown schematically above the sequences.