Figure 1.
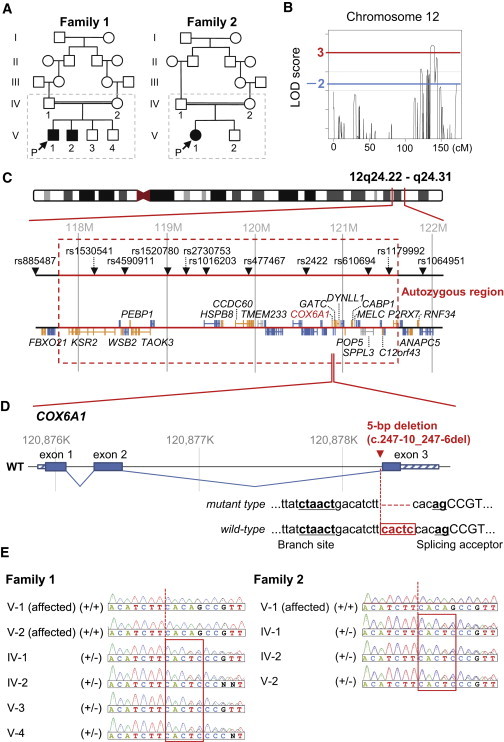
Genomic Analyses in the CMT Families
(A) Pedigrees of two consanguineous CMT families, from which a total of ten members in broken line boxes were sampled.
(B) Multipoint LOD scores on chromosome 12 using these ten members from two families with the HumanLinkage V SNP chip. We extracted genomic DNA from peripheral blood using the QIAamp DNA spin columns (QIAGEN) and quantified them using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies). Genotyping was performed with the GoldenGate Genotyping Universal-32 kit on a BeadArray Reader System (Illumina) according to the manufacturer’s assay guide. Genotype calls were made using the Genotyping module of the GenomeStudio v2009.1 software (Illumina). A multipoint linkage analysis was performed with Allegro v.24 and a human genetic map based on NCBI dbSNP Build 123 (see also Figure S1).
(C) Gene map in the significant linkage region on 12q24. These physical coordinates are taken from UCSC Genome Browser build hg19, RefSeq, and dbSNP 138 (UCSC Genome Browser database: 2014 update).
(D) A disease-specific 5 bp deletion (c.247−10_247−6delCACTC) in the pyrimidine tract near to the splicing acceptor near to the third exon of COX6A1.
(E) Validation and distribution of the 5 bp deletion among the CMT family members by Sanger-based PCR direct sequencing (see also Table S10).
