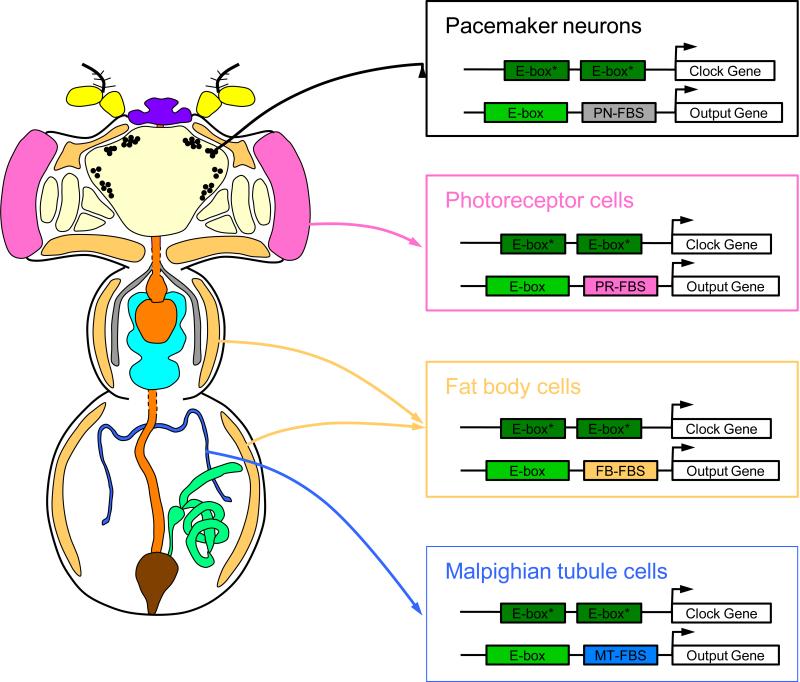
Figure 1.
Model for widespread and tissue-specific expression of CLK-CYC target genes. Clock gene-expressing tissues in an adult Drosophila (left). Yellow, antennae; cream, brain; tan, fat body; purple, proboscis; black, pacemaker neurons; pink, photoreceptors; orange, digestive tract; gray, salivary glands; aqua, ventral nerve chord; blue, Malpighian tubules; green, male reproductive tract; brown, rectum. Arrows denote regulation of clock gene and output gene expression in brain pacemaker neurons, photoreceptor cells, fat body cells and Malpighian tubules (right). A pair of closely-spaced E-boxes that bind CLK-CYC with high affinity are thought to promote transcription activation of clock genes in all tissues, whereas CLK-CYC bound to an E-box and a tissue-specific factor bound to a tissue-specific binding site cooperatively activate transcription of clock output genes in different tissues. Pacemaker neuron-specific factor binding site, PN-FBS; photoreceptor-specific factor binding site, PR-FBS; fat body-specific factor binding site, FB-FBS; Malpighian tubule-specific factor binding site, MT-FBS.