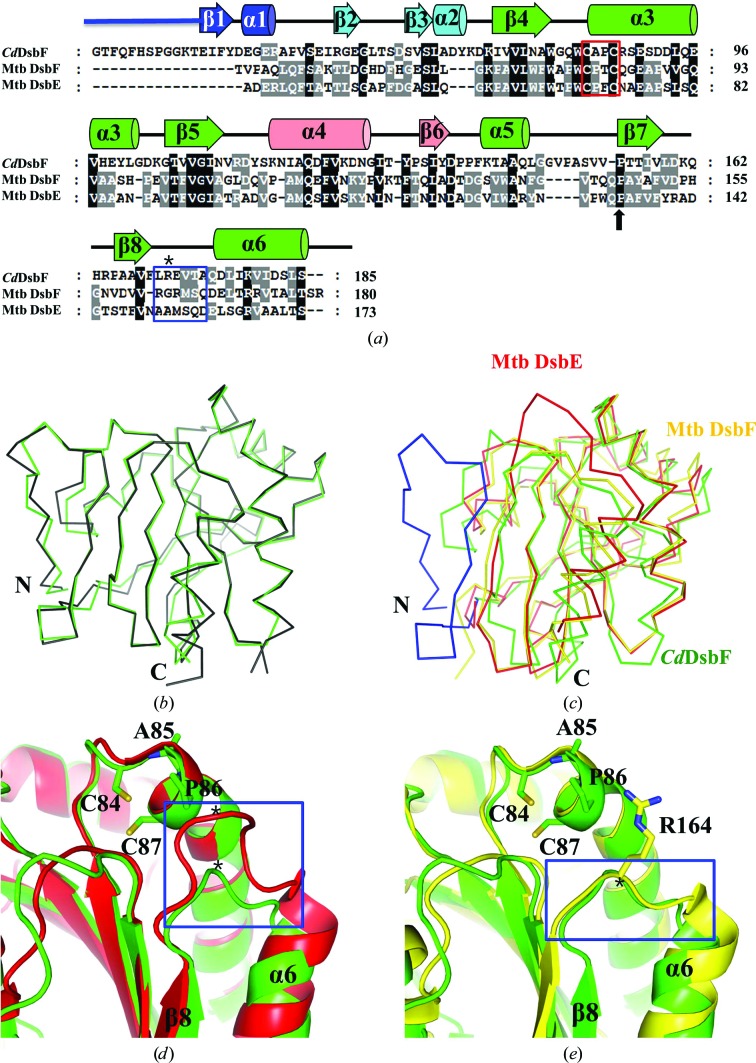
Figure 3.
Sequence and structural comparison with Mtb DsbE and Mtb DsbF. (a) Amino-acid sequence alignment of CdDsbF with Mtb DsbE and Mtb DsbF performed by ClustalX and modified manually based on the structure-based alignment. Secondary-structure elements are shown based on the structure of CdDsbF. The CXXC motif and cis-Pro are indicated by a red box and a black arrow, respectively. The β8–α6 loop indicated by a blue box and the residues marked with asterisks are mentioned in the main text and in (d) and (e). (b) Superimposition of the structures of CdDsbF (green) and C. glutamicum thioredoxin-like protein (PDB entry 3lwa; black). (c) Superimposition of the structures of CdDsbF (green), Mtb DsbE (red) and Mtb DsbF (yellow). CdDsbF has a longer N-terminal region (blue) compared with Mtb DsbE and Mtb DsbF. (d) Structure superimposition of a loop region of CdDsbF (green) and Mtb DsbE (red). The loop region (blue box) connecting β8 to α6 is closer to the CXXC motif in Mtb DsbE than in CdDsbF in the three-dimensional structures. (e) Structure superimposition of a loop region of CdDsbF (green) and Mtb DsbF (yellow). The loop region (blue box) connecting β8 to α6 is at a similar distance to the CXXC motif in the three-dimensional structures.