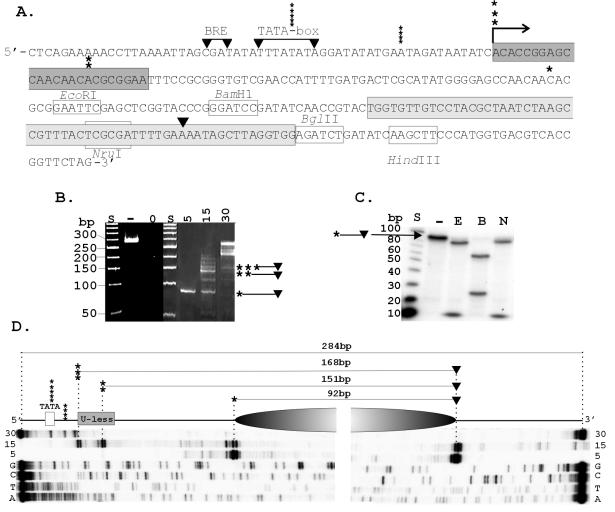
FIG. 1.
Transcription template and position of HMtA2 assembly. (A) The sequence of the transcription template is shown with the BRE and TATA-box sequence from the HMtB promoter (16), the U-less cassette (dark gray), Selex1 sequence (light gray) (4), site of transcription initiation (→), and 3′ (▾) and 5′ (asterisks) boundaries of the MN-protected fragments indicated. (B) Electrophoretic separation of the fragments of the template DNA protected from MN digestion by assembly into complexes at the HMtA2 dimer/100-bp ratios indicated above the corresponding lane. Control lanes contained size standards (S), untreated template DNA (−) and template DNA exposed to MN for 1 min in the absence of HMtA2 (0). (C) Autoradiogram of the electrophoretic separation of the restriction fragments generated from a population of the ∼90-bp MN-protected DNA fragments after 32P end labeling and digestion with EcoRI (E), BamHI (B), or NruI (N). Control lanes contained size standards (S) and an aliquot of the ∼90-bp molecules not exposed to restriction enzymes (−). (D) Diagram showing the position of the archaeal nucleosome (shaded oval), the 5′ and 3′ boundaries, and the precise lengths of the fragments of the template DNA protected from MN digestion as determined by the lengths of the primer extension products indicated below. The extension products were generated from 32P-labeled primers that hybridized to the MN-protected fragments bound by 5, 15, or 30 HMtA2 dimers per 100 bp of template DNA. Adjacent lanes contained the products of sequencing reactions (G, C, T, A) generated from the template DNA, using the same primers.