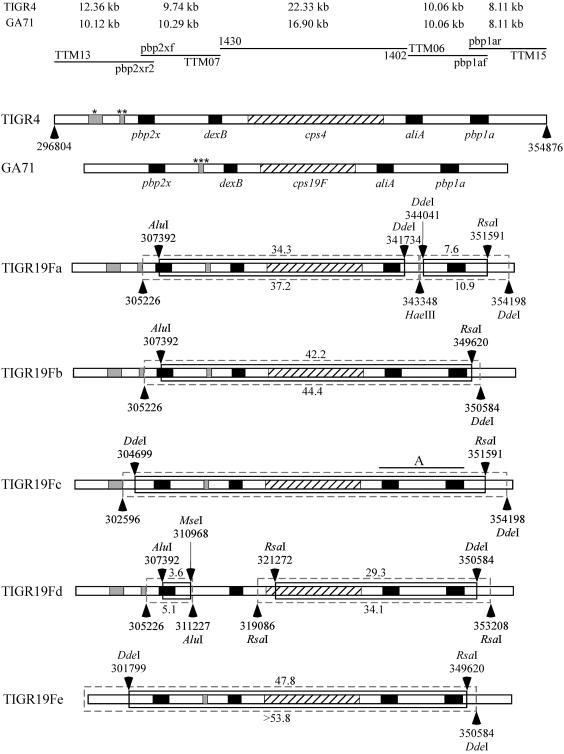
FIG. 1.
Graphic representations of the recombinational replacements within and outside the pbp2x-cps-pbp1a region identified by RFLP analysis in five isolates of TIGR4 transformed independently to penicillin nonsusceptibility and serotype 19F with chromosomal DNA of strain GA71. Horizontal lines at the top represent products of PCR amplified with the oligonucleotides indicated above (forward primer) and below (reverse primer) the line. See Materials and Methods for detailed descriptions of the primers used. The sizes of the amplicons generated for the recipient (TIGR4) and donor (GA71) strains with a particular pair of primers are shown above the lines representing PCR products. Horizontal bars represent regions of genomic DNA in the strains analyzed (indicated on the left). The cross-hatched regions represent cps loci, and the black rectangles depict genes present in the regions flanking the cps locus in all natural isolates of S. pneumoniae. Gray regions indicate indels located outside the dexB-aliA locus. The possible sites of the indels were identified on the basis of the Spain23F-1 clone (serotype 23F isolate genome sequences were made available by the Sanger Institute [http://www.sanger.ac.uk/Projects/S_pneumoniae]). The top bar presents the genetic organization of the 58.1-kbp fragment of the TIGR4 strain from position 296804 to position 354876 of its published genome sequence (GenBank accession number NC003028) (40). The single star marks a 1,709-bp indel at position 300888 to position 302596, and the double star marks a 553-bp indel at position 304674 to position 305226, both upstream of pbp2x in TIGR4. The second horizontal bar represents the organization of the same region in isolate GA71, with a size of 51.0 kbp, calculated on the basis of the PCR products generated in this study. The triple star marks the 559-bp indel present in the Sanger Spain23F-1 clone strain at position 3809 bp downstream of pbp2x, also identified in GA71. Arrows above and below bars point at the restriction sites identified as the limits of possible crossover, as described in Materials and Methods. Lack of an enzyme name next to the restriction site genome position number indicates that the crossover site was eliminated by the indel. Inner, solid boxes represent the minimal replacements identified by RFLP analysis in capsule and PBP transformants of TIGR4. Estimated minimal TIGR4 genomic DNA fragments involved in recombinational replacements are depicted above boxes. Outer, dashed boxes represent maximal replacements identified the same way, and the estimated sizes of maximal replacements are depicted below boxes. Fingerprints generated for isolates TIGR19Fa, TIGR19Fc, and TIGR19Fd indicate the presence of two recombinational replacements. The RFLP technique used did not allow us to narrow the sizes of replacements in TIGR19Fc. Region A, extending from a position 611 bp upstream of aliA to one 159 bp downstream of pbp1a, marks the possible site of the additional recombinational crossover(s) in this transformant. Because of the presence of these additional crossovers, the sizes of the replacements were not calculated.