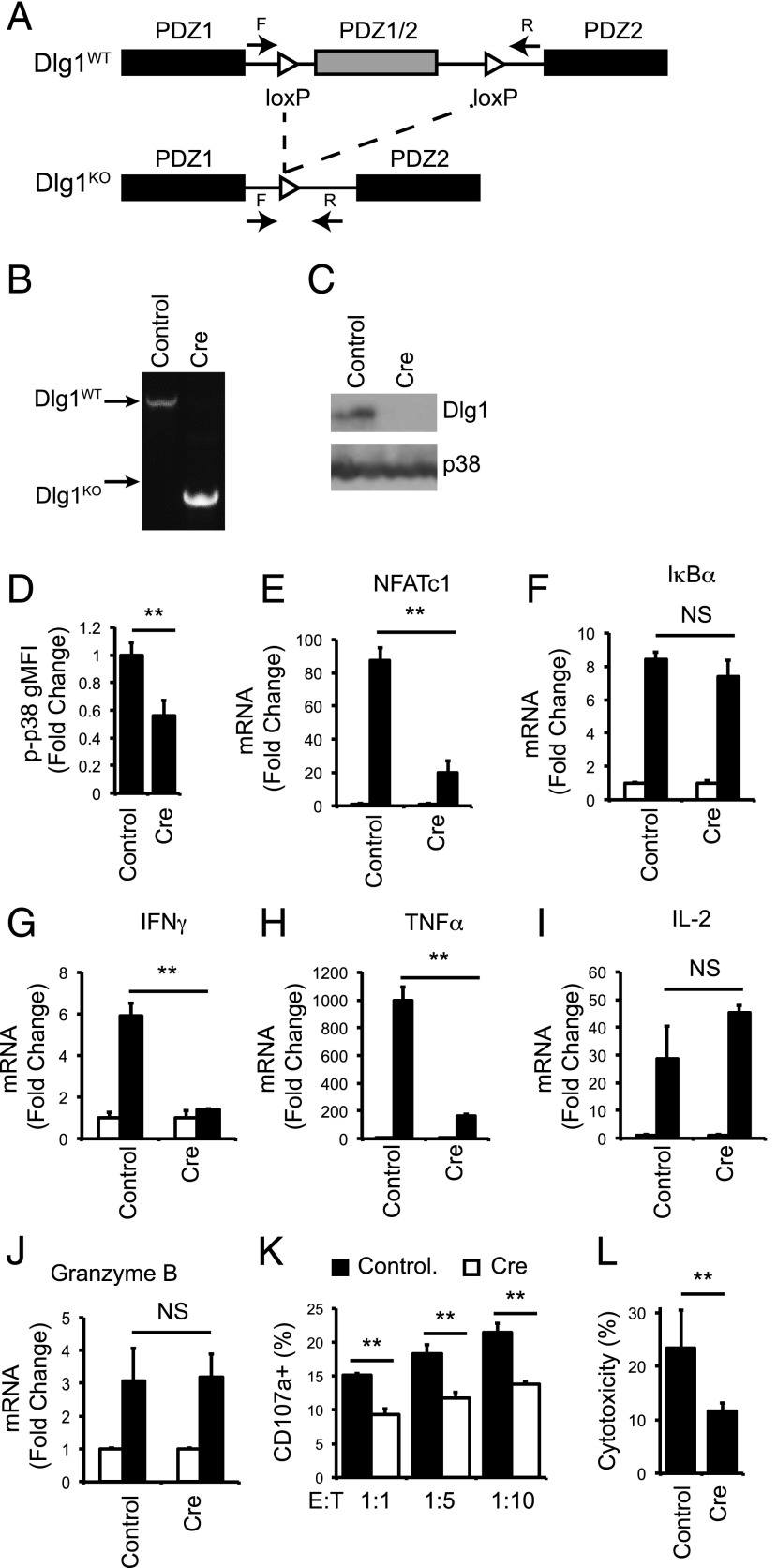
FIGURE 6.
Acute knockout of Dlg1 impairs p38 activation and NFAT-dependent proinflammatory cytokines gene expression and target cell lysis in response to TCR stimulation. (A) Genomic organization of Dlg1flox/flox mice as described previously (28). F and R refer to the location of the forward and reverse primers for gDNA analysis. (B–L) CD8+ T cells were isolated from spleens of OT-1 Dlg1flox/flox mice and expanded on plate-bound anti-CD3 and anti-CD28 Ab for 48–72 h, followed by infection with retroviral cre recombinase or vector control. (B) Genomic DNA was isolated, and PCR analysis was performed. (C) Whole-cell lysates were subjected to SDS-PAGE and blotted with anti-Dlg1 or anti-p38. (D) Cells were stimulated with plate-bound anti-CD3 and anti-CD28 Ab for 15 min, followed by fixation, permeabilization, and staining for p-p38 180/182. Error bars represent SD of three independent experiments. (E–J) Cells were left unstimulated or stimulated with plate-bound anti-CD3 and anti-CD28 for 2 h (E and F) or 6 h (G–J), followed by mRNA isolation, reverse transcription, and qPCR analysis using primers specific for NFATc1(E), IκBα (F), IFN-γ (G), TNF-α (H), IL-2 (I), or granzyme B (J). All values were normalized to L32 and unstimulated values set to 1.0. Error bars represent stand deviation of triplicates; data are representative of four independent experiments. (K and L) Cells were incubated with EG.7 thymoma cells at indicated ratios for 2 h, following surface staining for CD107a (K) or lactate dehydrogenase cytotoxicity assay (L). **p < 0.05.