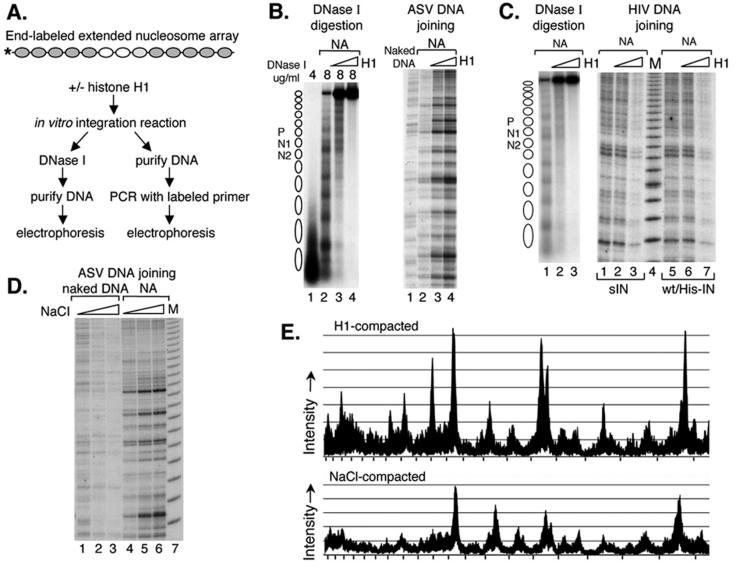
FIG. 3.
Influence of target nucleosome array compaction on integrase-mediated joining of viral DNAs. (A) Experimental design. The extended nucleosome array was incubated with increasing amounts of histone H1 for 1 h, followed by addition of the ASV or HIV-1 integration components. One hour later, the mixture was analyzed by DNase I assay and by PCR with the N2-specific target DNA primer and an HIV-1 or ASV donor DNA primer. (B) DNase I analysis of the target DNA after incubation with the ASV DNA joining components (on the left) and PCR detection of the ASV DNA joining pattern (on the right). NA, extended nucleosome array. On the left side, DNase I concentrations are shown above the gel. Right side: lane 1, naked DNA target; lane 2, extended nucleosome array without histone H1; lane 3, array compacted by addition of 7.5 nM histone H1; lane 4, extended array compacted by addition of 13 nM histone H1. (C) DNase I analysis of the DNA used as a target for HIV-1 DNA joining (on the left) and PCR detection of the HIV-1 DNA joining pattern (on the right). DNase I concentrations used (left side, 4 μg/ml for lane 1 and 8 μg/ml for lanes 2 and 3). Right side: lanes 1 to 3, HIV-1sIN-mediated DNA joining; lanes 5 to 7, HIV-1wt/His IN-mediated DNA joining; lanes 1 and 5, extended nucleosome array; lanes 2 and 6, extended array compacted by addition of 7.5 nM histone H1; lanes 3 and 7, extended array compacted by addition of 13 nM histone H1; lane 4, 10-bp molecular weight marker (M). (D) ASV integrase-mediated joining into NaCl-mediated compacted targets. Naked DNA or the extended nucleosome array was incubated with 100 mM NaCl (lanes 2 and 5, respectively) or 120 mM NaCl (lanes 3 and 6, respectively) and then incubated with the ASV integration components. Lanes 1 and 4, ASV DNA joining to naked DNA and the extended nucleosome array in the presence of control buffer; lane 7, 10-bp molecular weight marker (M). (E) Comparison of ASV integration patterns in the region of nucleosome N1 with extended and compacted nucleosomal arrays. The sequencing gels of panels B, right side, lanes 2 and 4 (top tracing), and D, lanes 4 and 6 (bottom tracing), were scanned with a Fuji BAS-250 Bio-imaging analyzer and analyzed with Image Gauge 4.0 software. Tick marks on the x axis correspond to the 10-bp DNA molecular weight marker.