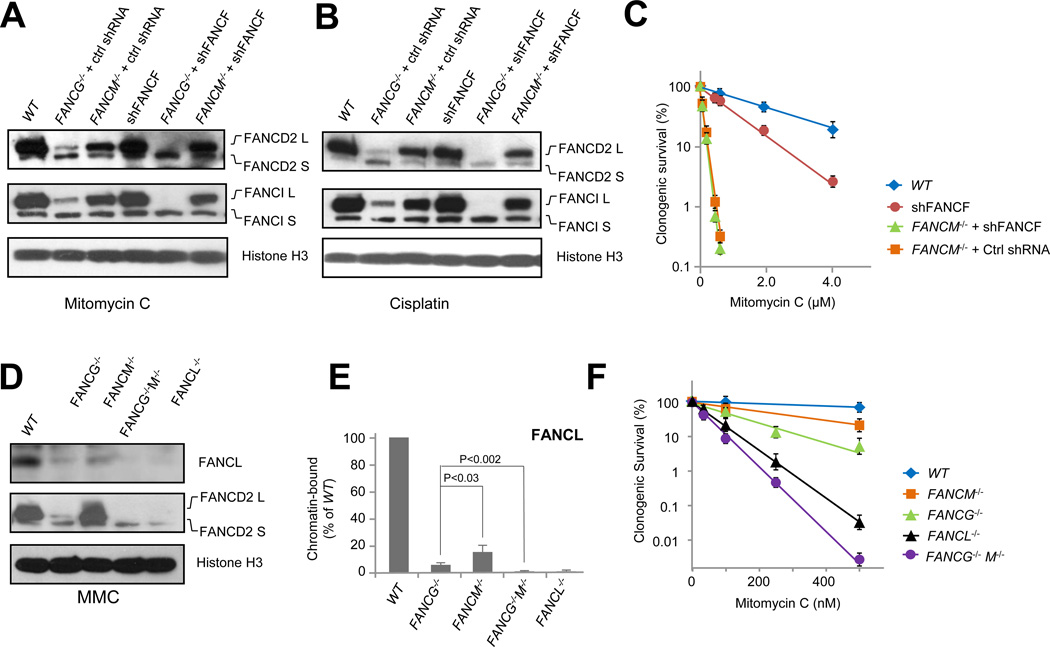
Fig. 4. FANCM functions epistatically with the C-E-F module in FA core complex chromatin association.
(A) Immunoblot detecting mitomycin C-induced monoubiquitination of FANCD2 and FANCI in wild-type HCT116 cells (WT) and the indicated knockout/knockdown mutant cells. See also Fig. S4.
(B) Immunoblot detecting cisplatin-induced monoubiquitination of FANCD2 in wild-type HCT116 cells (WT) and the indicated knockout/knockdown mutant cells. See also Fig. S4.
(C) Clonogenic survival of HCT116 FANCF knockdown alone (shFANCF), FANCM−/− +shFANCF, and FANCM−/− +Ctrl shRNA mutants treated with mitomycin C. See also Fig. S4.
(D) Immunoblot detecting MMC-induced monoubiquitination of FANCD2 and chromatin binding of FANCL in wild-type HCT116 cells (WT) and the indicated mutant cells exposed to mitomycin C. Histone H3 serves as a loading control for chromatin-bound protein fractions. See also Fig. S2F.
(E) Quantification of (C) - chromatin-bound FANCL in FANCG−/−, FANCM−/−, and FANCG−/− M−/− double mutants. See also Fig. S2F.
(F) Clonogenic survival of wild-type HCT116 cells (WT), FANCM−/−, FANCG−/−, FANCL−/−, and FANCG−/− M−/− mutants treated with mitomycin C. See also Fig. S2F.
Error bars for chromatin fraction represent SDs derived from three independent experiments. Error bars for clonogenic survival are derived from SDs from four ndependent tests done in triplications.