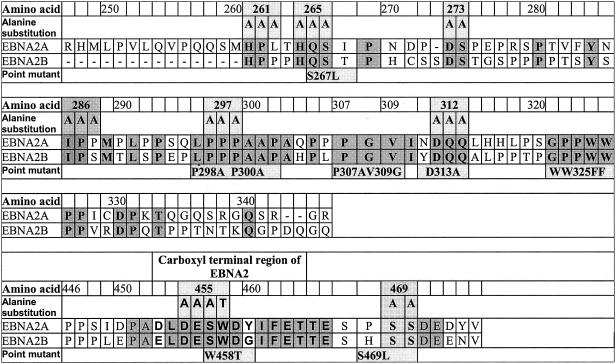
FIG. 1.
Mutant alleles of EBNA2 used in this study. Portions (amino acids 247 to 346 and 446 to 475) of the derived amino acid sequence of the coding sequence of the EBNA2A gene from the M-ABA strain of EBV that encompasses the divergent and carboxyl-terminal regions are depicted. For comparison, the sequence encoded by the EBNA2B allele (8) is indicated. Amino acid identities between A and B types are shaded. Point mutations that were constructed for this study are indicated below the sequence, and alanine substitution mutations are depicted above the sequence. Alanine substitution mutations were generated by standard techniques, using PCR and oligonucleotides that introduced the alanine substitution mutations indicated (19). The modification resulting in the alanine substitutions also generated a new PstI site within the coding region in addition to a preexisting PstI site that resides at its 3′ end. Specific PCR-generated fragments representing the amino and carboxyl portions of these mutations were cleaved with appropriate enzymes and ligated together in the pSG5 expression vector. All mutants were sequenced to verify the presence of the mutation and the absence of additional mutations. pSG5FLAG-Ini1 was constructed by introducing two tandem FLAG-encoding sequences at the amino terminus of the entire hSFN5/Ini1 open reading frame in the pSG5 vector.