Fig. 1.
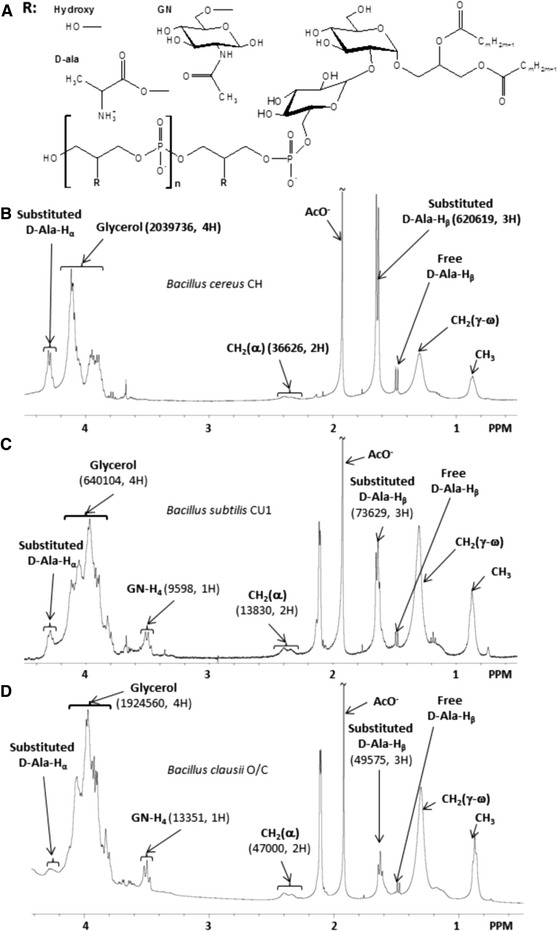
(a) General model of the molecular structure of Bacillus LTAs as determined from 1H NMR analysis. n represents the number of glycerol-phosphate repeating units of the hydrophilic backbone, and R the substituent group of glycerol. R can be hydroxyl groups (Hydroxy), d-alanine (d-Ala) or N-acetylglucosamine (GN). (b) 1H NMR spectrum of B. cereus CH LTA. (c) 1H NMR spectrum of B. subtilis CU1 LTA. (d) 1H NMR spectrum of B. clausii O/C LTA. Areas (in a.u.) and the corresponding number of hydrogen used for chain length, proportion of glycerol esterified with alanine and proportion of glycerol substituted with N-acetylglucosamine calculations, are indicated in bracket
