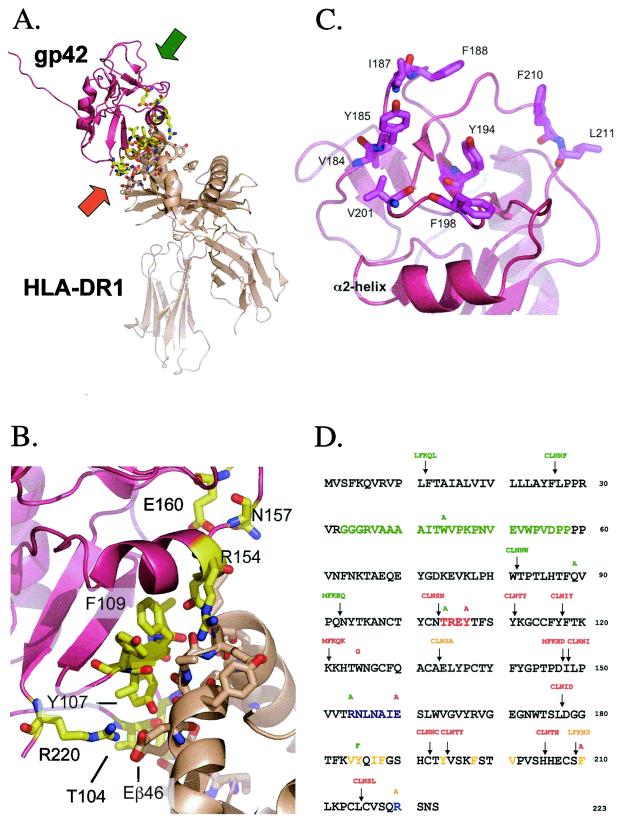
FIG. 1.
Important structural features of EBV gp42 and residue sequence mapping location of mutations. (A) Three-dimensional ribbon model of soluble EBV gp42 (rose) bound to class II HLA-DR1 (beige). The amino terminus of residues 33 to 85 was disordered in the crystal structure. Backbones of residues interacting with HLA class II are yellow, oxygen atoms are red, and nitrogen atoms are blue. The orange and green arrows indicate the regions enlarged in panels B and C, respectively. (B) Close-up depicting key gp42 contact residues at the HLA class II interface. The color scheme is the same as in panel A. (C) Close-up of gp42 hydrophobic pocket highlighting aliphatic and aromatic residues. (D) The residue sequence of gp42 reveals the locations of point mutations above residues and linker insertion mutations containing 5-amino-acid inserts above arrows. Linker insertion mutants are named based on the residue that their mutation precedes, e.g., LI12, and point mutants reveal original and mutation residue, e.g., W44A. The baculovirus-produced soluble gp42 (sgp42) used as a positive control in many experiments spans residues 33 through 223 and was kindly provided by Maureen Mullen. Residues of the potential gH/gL binding site are in green. HLA class II contact sites are indicated as follows: aromatic ring residues in red, α-2-helix in purple, and arginine 220 in blue. Hydrophobic residues in the pocket are in yellow. Mutants are color-coded by their function in the fusion assay: those in green have levels similar to the wild-type levels, those in orange have reduced levels of fusion, and those in red do not mediate fusion.