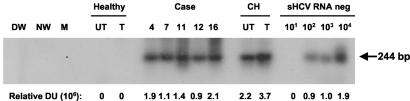
FIG. 6.
Detection of the HCV RNA negative strand in PBMC from individuals with clinical and apparently complete virological resolution of hepatitis C. Total RNA was extracted from IL-2- and PHA-stimulated PBMC isolated from individuals with self-limited (case 4) or therapeutically induced (cases 7, 11, 12, and 16) recovery from hepatitis and from control untreated (UT) and IL-2- and PHA-treated (T) PBMC obtained from a healthy donor and a patient with actively progressing chronic hepatitis C (CH). RNA was reverse transcribed and cDNA was amplified by PCR with HCV RNA negative strand-specific 5′-UTR primers. Serial dilutions of a synthetic HCV RNA negative strand (sHCV RNA neg) amplified in parallel were used as semiquantitative standards. Water (DW and NW) and mock (M) samples were used as negative and contamination controls. The positive signals (244-bp fragments) were visualized by hybridization to the recombinant HCV UTR-E2 probe. Values under the panel represent relative densitometric units (DU) given by positive hybridization signals.