Figure 1.
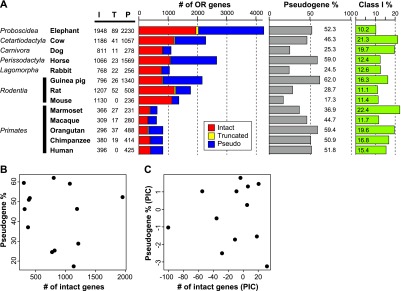
Numbers of OR genes in the genome sequence from 13 placental mammal species. (A) “I,” “T,” and “P” represent the number of intact genes, truncated genes, and pseudogenes, respectively. An intact gene was defined as a sequence starting from an initiation codon and ending with a stop codon that did not contain any disrupting mutations. A pseudogene was defined as a sequence with a nonsense mutation, frameshift, deletion within conserved regions, or some combination thereof. A truncated gene was defined as a partial, intact sequence located at a contig end. An intact gene was assumed to be functional, while a truncated gene was presumed to be either a functional gene or a pseudogene. The fraction of OR pseudogenes was calculated as the number of OR pseudogenes divided by the total number of OR genes. The fraction of Class I genes was calculated as the number of intact Class I genes divided by the total number of intact OR genes. Dog and rat data were taken from Niimura and Nei (2007), and the data for the five primates were from Matsui et al. (2010). (B) There was no significant correlation between the number of intact OR genes within a genome and the fraction of OR pseudogenes within that same genome among these 13 species (r = −0.137; P = 0.655); (C) again, there was no significant correlation (r = −0.003; P = 0.992) after the comparative method of phylogenetically independent constants (PICs) was used to remove phylogenetic dependence (Felsenstein 1985).
