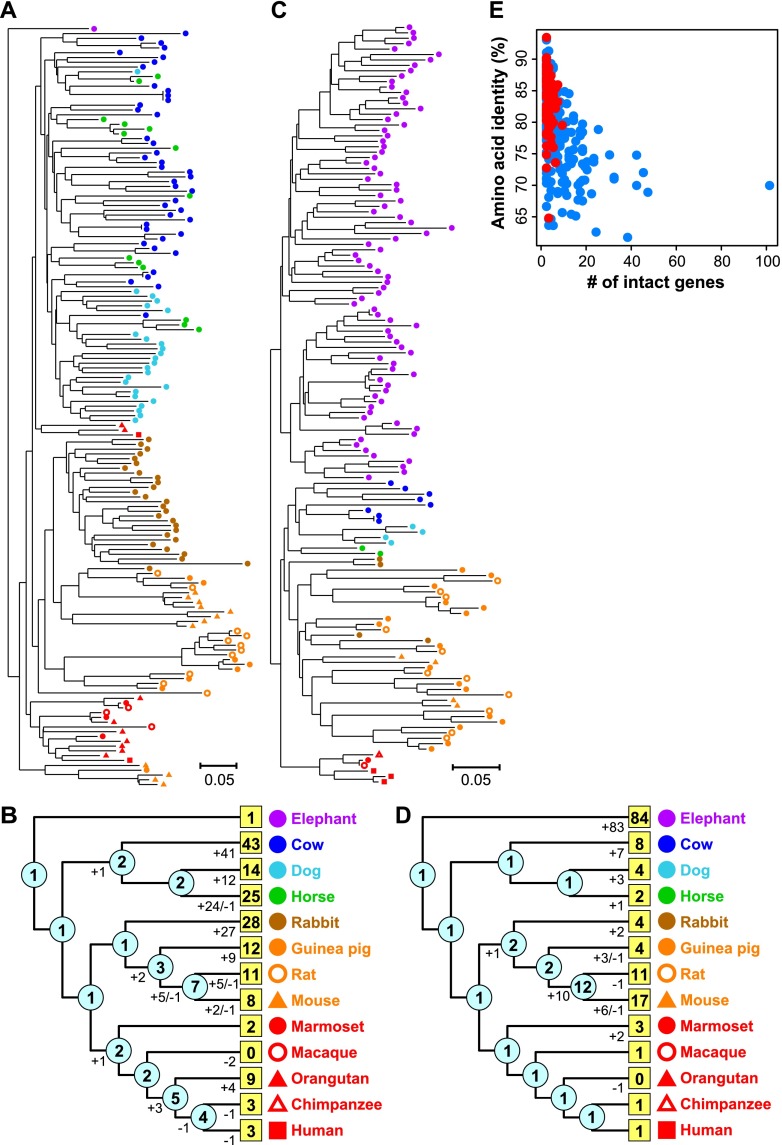
Figure 3.
Expanded OGGs. Neighbor-joining (NJ) phylogenetic trees were constructed from all intact OR genes in OGG2-1 (A) and OGG2-2 (C). In each tree (A,C), a colored symbol indicates a gene from the species depicted in B and D. Each scale bar indicates the number of amino acid substitutions per site. Gene names and bootstrap values are shown in Supplemental Figure S5A,B. (B,D) Number of gene gains and losses in each branch and the number of genes at ancestral nodes (shown in cyan circles) calculated from the OGG2-1 (A) and OGG2-2 (C) trees, respectively, by the reconciled-tree method (Niimura and Nei 2007). A number in a yellow box indicates the number of intact OR genes in each species belonging to each OGG. (E) For each OGG, the total number of intact genes in elephant and mouse within respective OGGs was negatively correlated with amino acid sequence identity between elephant and mouse among intact OR genes within the respective OGGs (rs = −0.52; P < 2.2 × 10−16). In all, 414 OGGs that contained at least one intact gene from both elephant and mouse were considered. When an OGG included two or more genes from either or both species, the mean of the amino acid sequence identities for all possible interspecies combinations of genes was used.